nf-core/phageannotator
Pipeline for identifying, annotation, and quantifying phage sequences in (meta)-genomic sequences.
Introduction
nf-core/phageannotator is a bioinformatics pipeline for identifying, annotation, and quantifying phage sequences in (meta)-genomic sequences.
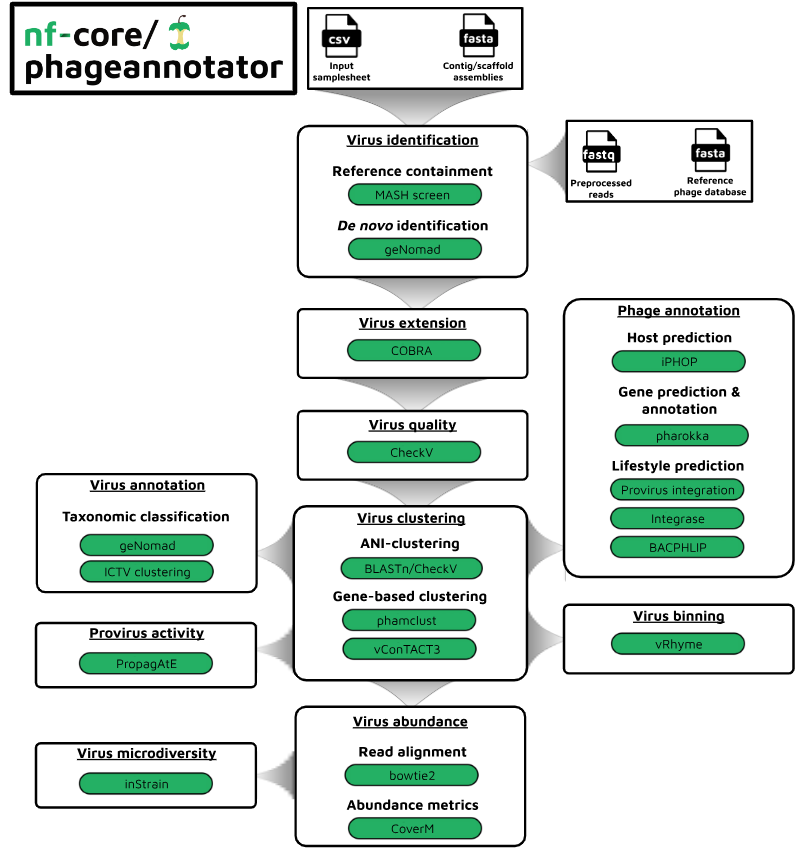
Pipeline summary
The core identification/quantification portion of this pipeline takes (meta)-genomic assemblies (as output by nf-core/mag) and performs the following steps:
- Phage sequence identification
- Quality assessment/filtering (
CheckV) - ANI clustering/dereplication (
BLAST)(CheckV) - Binning (
VRhyme) - Abundance estimation (
bowtie2)(CoverM)- Output can be used as input for nf-core/differentialabundance
- OPTIONAL - Strain profiling (
inStrain) - OPTIONAL - Prophage activity prediction (
propagAtE)
The annotation portion of this pipeline performs the following steps:
- Marker-based taxonomic classification (
geNomad) - Genome proximity taxonomic classification (
mash) - Host prediction (
iPHoP) - Lifestyle prediction (
BACPHLIP) - Protein-coding gene prediction (
Prodigal-gv)- Output can be used as input for nf-core/funcscan
Quick Start
-
Install
Nextflow(>=22.10.1). -
Install any of
Docker,Singularity(you can follow this tutorial),Podman,ShifterorCharliecloudfor full pipeline reproducibility (you can useCondaboth to install Nextflow itself and also to manage software within pipelines. Please only use it within pipelines as a last resort; see docs). -
Download the pipeline and test it on a minimal dataset with a single command:
nextflow run nf-core/phageannotater -profile test,YOURPROFILE --outdir <OUTDIR>Note that some form of configuration will be needed so that Nextflow knows how to fetch the required software. This is usually done in the form of a config profile (
YOURPROFILEin the example command above). You can chain multiple config profiles in a comma-separated string.- The pipeline comes with config profiles called
docker,singularity,podman,shifter,charliecloudandcondawhich instruct the pipeline to use the named tool for software management. For example,-profile test,docker. - Please check nf-core/configs to see if a custom config file to run nf-core pipelines already exists for your Institute. If so, you can simply use
-profile <institute>in your command. This will enable eitherdockerorsingularityand set the appropriate execution settings for your local compute environment. - If you are using
singularity, please use thenf-core downloadcommand to download images first, before running the pipeline. Setting theNXF_SINGULARITY_CACHEDIRorsingularity.cacheDirNextflow options enables you to store and re-use the images from a central location for future pipeline runs. - If you are using
conda, it is highly recommended to use theNXF_CONDA_CACHEDIRorconda.cacheDirsettings to store the environments in a central location for future pipeline runs.
- The pipeline comes with config profiles called
-
Start running your own analysis!
nextflow run nf-core/phageannotater --input samplesheet.csv --outdir <OUTDIR> -profile <docker/singularity/podman/shifter/charliecloud/conda/institute>nf-core/phageannotator is a bioinformatics pipeline that …
Usage
If you are new to Nextflow and nf-core, please refer to this page on how to set-up Nextflow. Make sure to test your setup with -profile test before running the workflow on actual data.
Now, you can run the pipeline using:
nextflow run nf-core/phageannotator \
-profile <docker/singularity/.../institute> \
--input samplesheet.csv \
--outdir <OUTDIR>Please provide pipeline parameters via the CLI or Nextflow -params-file option. Custom config files including those provided by the -c Nextflow option can be used to provide any configuration except for parameters;
see docs.
For more details and further functionality, please refer to the usage documentation and the parameter documentation.
Documentation
The nf-core/phageannotater pipeline comes with documentation about the pipeline usage, parameters and output.
Credits
nf-core/phageannotator was originally written by @CarsonJM.
We thank the following people for their extensive assistance in the development of this pipeline:
Contributions and Support
If you would like to contribute to this pipeline, please see the contributing guidelines.
For further information or help, don’t hesitate to get in touch on the Slack #phageannotator channel (you can join with this invite).
Citations
An extensive list of references for the tools used by the pipeline can be found in the CITATIONS.md file.
You can cite the nf-core publication as follows:
The nf-core framework for community-curated bioinformatics pipelines.
Philip Ewels, Alexander Peltzer, Sven Fillinger, Harshil Patel, Johannes Alneberg, Andreas Wilm, Maxime Ulysse Garcia, Paolo Di Tommaso & Sven Nahnsen.
Nat Biotechnol. 2020 Feb 13. doi: 10.1038/s41587-020-0439-x.