nf-core/proteinannotator
Generation of sequence-level annotations for amino acid sequences
Introduction
nf-core/proteinannotator is a bioinformatics pipeline that computes statistics for protein FASTA inputs and produces protein annotations based on predicted sequence features, including conserved domains, functions, and secondary structure.
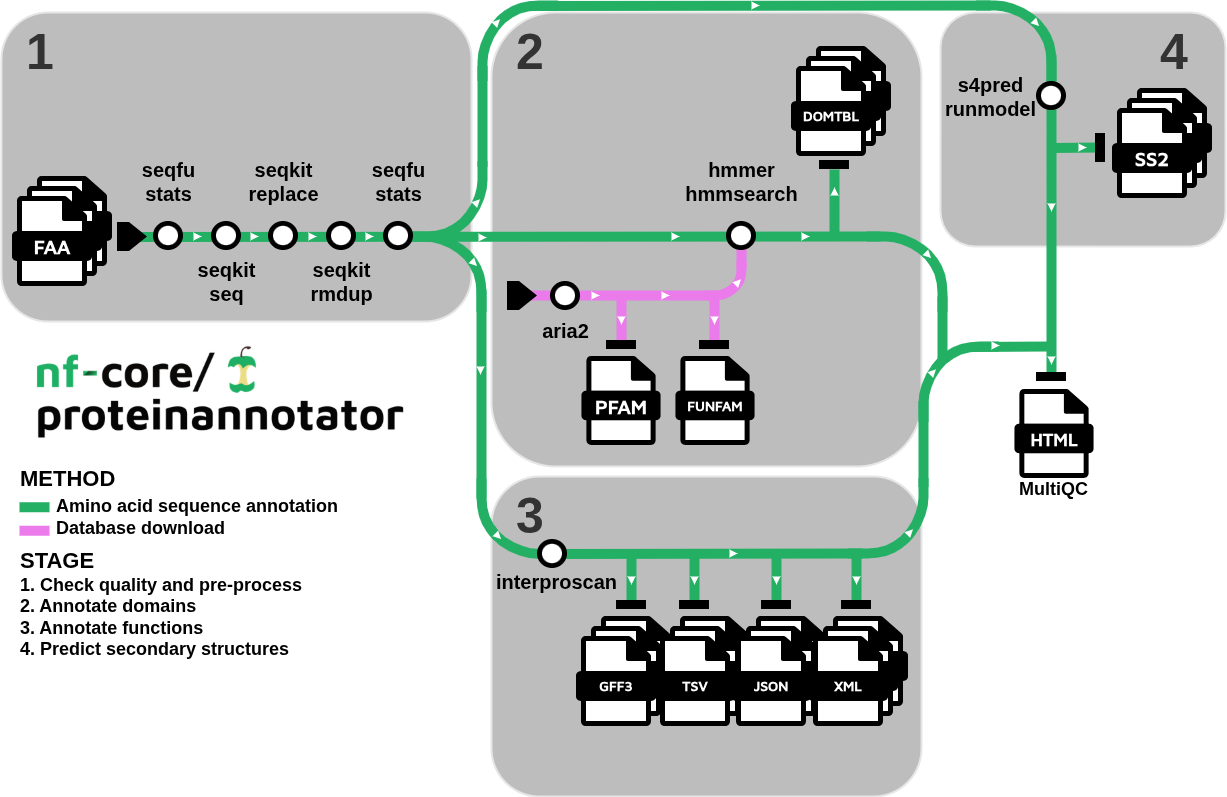
Check quality and pre-process
Generate input amino acid sequence statistics with (SeqFu) and pre-process them (i.e., gap removal, convert to upper case, validate, filter by length, replace special characters such as /, and remove duplicate sequences) with (SeqKit)
Annotate sequences
- Conserved domain annotation with (
hmmer) against databases such as Pfam and FunFam - Functional annotation:
- (
InterProScan) a software tool used to analyze protein sequences by scanning them against the signatures of protein families, domains, and sites in the InterPro database, helping to identify their functional characteristics.
- (
- Predict secondary structure compositional features such as α-helices, β-strands and coils with (
s4pred) - Present QC stats for input sequences before and after initial pre-processing with (
MultiQC)
Usage
If you are new to Nextflow and nf-core, please refer to this page on how to set-up Nextflow. Make sure to test your setup with -profile test before running the workflow on actual data.
First, prepare a samplesheet with your input data that looks as follows:
samplesheet.csv:
id,fasta
species1,species1_proteins.fasta
species2,species2_proteins.fastaEach row represents a FASTA file of proteins from a single species.
Now, you can run the pipeline using:
nextflow run nf-core/proteinannotator \
-profile <docker/singularity/.../institute> \
--input samplesheet.csv \
--outdir <OUTDIR>Please provide pipeline parameters via the CLI or Nextflow -params-file option. Custom config files including those provided by the -c Nextflow option can be used to provide any configuration except for parameters; see docs.
For more details and further functionality, please refer to the usage documentation and the parameter documentation.
Pipeline output
To see the results of an example test run with a full size dataset refer to the results tab on the nf-core website pipeline page. For more details about the output files and reports, please refer to the output documentation.
Credits
nf-core/proteinannotator was originally written by Olga Botvinnik and Evangelos Karatzas.
We thank the following people for their extensive assistance in the development of this pipeline:
Contributions and Support
If you would like to contribute to this pipeline, please see the contributing guidelines.
For further information or help, don’t hesitate to get in touch on the Slack #proteinannotator channel (you can join with this invite).
Citations
An extensive list of references for the tools used by the pipeline can be found in the CITATIONS.md file.
You can cite the nf-core publication as follows:
The nf-core framework for community-curated bioinformatics pipelines.
Philip Ewels, Alexander Peltzer, Sven Fillinger, Harshil Patel, Johannes Alneberg, Andreas Wilm, Maxime Ulysse Garcia, Paolo Di Tommaso & Sven Nahnsen.
Nat Biotechnol. 2020 Feb 13. doi: 10.1038/s41587-020-0439-x.