nf-core/ribomsqc
QC pipeline that monitors mass spectrometer performance in ribonucleoside analysis

Introduction
Use nf-core/ribomsqc to:
- Perform automated quality control of ribonucleoside analysis by mass spectrometry.
- Summarize and visualize QC metrics through integrated MultiQC reports.
 Figure 1: General workflow overview showing the pipeline steps from RAW file input through ThermoRawFileParser conversion, MSNBase XIC extraction, JSON merging, to final MultiQC report generation.
Figure 1: General workflow overview showing the pipeline steps from RAW file input through ThermoRawFileParser conversion, MSNBase XIC extraction, JSON merging, to final MultiQC report generation.
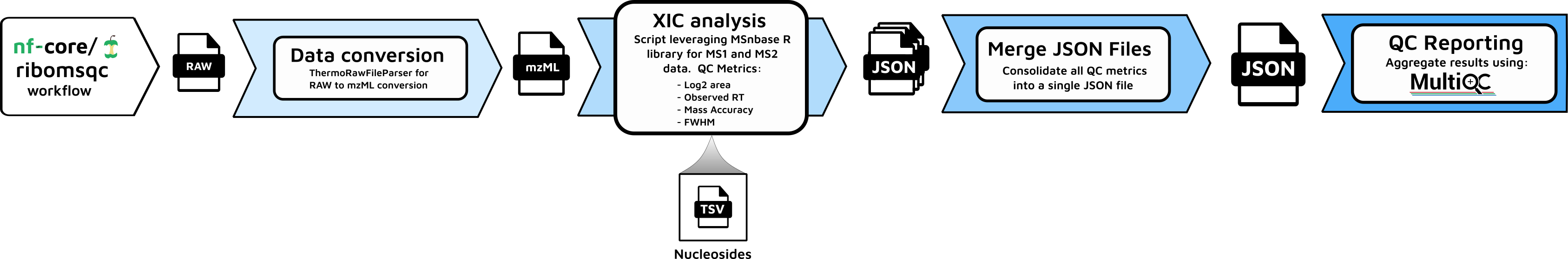 Figure 2: Detailed workflow diagram illustrating the data flow and process connections in the ribomsqc pipeline.
Figure 2: Detailed workflow diagram illustrating the data flow and process connections in the ribomsqc pipeline.
Usage
If you are new to Nextflow and nf-core, please refer to this page on how to set-up Nextflow.
-
Prepare a samplesheet with your input data, for example:
samplesheet.csv id,raw_file Day_5,path/to/Day_5.raw
For more information, see the usage docs on required samplesheet.csv columns.
- Prepare an analytes TSV file (e.g.
qcn1.tsv) with your compounds and theoretical retention times. The TSV must have exactly these columns and format:
short_name long_name mz_M0 mz_M1 mz_M2 ms2_mz rt_teoretical
C Cytidine 50 μg/mL 244.0928 112.0505 555
U Uridine 25 μg/mL 245.0768 113.0346 1566
m3C 3-Methylcytidine methosulfate 100 μg/mL 258.1084 126.0662 508
m5C 5-Methylcytidine 100 μg/mL 258.1084 126.0662 655
Cm 2-O-Methylcytidine 20 μg/mL 258.1084 112.0505 883
m5U 5-Methyluridine 50 μg/mL 259.0925 127.0502 1866
I Inosine 25 μg/mL 269.088 137.0458 1741
m1A 1-Methyladenosine 25 μg/mL 282.1197 150.0774 523
G Guanosine 25 μg/mL 284.0989 152.0567 1726
m7G 7-Methylguanosine 25 μg/mL 298.1146 166.0723 554Replace only the values in the rt_teoretical column with your own retention times (in seconds) for each compound.
-
Run the pipeline:
nextflow run nf-core/ribomsqc \ --input samplesheet.csv \ --analytes_tsv qcn1.tsv \ --analyte all \ --rt_tolerance 120 \ --mz_tolerance 7 \ --ms_level 1 \ --outdir results \ -profile singularity
Please provide pipeline parameters via the CLI or Nextflow -params-file option.
For more information, see the usage docs and parameters.
Pipeline output
See results page for example output and output docs.
Credits
nf-core/ribomsqc was originally written by Roger Olivella.
Contributions and Support
For help, visit Slack #ribomsqc or see contributing guide.
Citations
See CITATIONS.md for tool references.
Ewels PA et al. (2020) The nf-core framework. Nat Biotechnol. doi:10.1038/s41587-020-0439-x












