nf-core/smartseq2
A pipeline for processing single cell RNA-seq data generated with the SmartSeq2 protocol.
This pipeline is archived and no longer maintained.
Archived pipelines can still be used, but may be outdated and will not receive bugfixes.
22.10.6.
Learn more.
Introduction
The pipeline is built using Nextflow, a workflow tool to run tasks across multiple compute infrastructures in a very portable manner. It comes with docker containers making installation trivial and results highly reproducible.
Quick Start
i. Install nextflow
ii. Install either Docker or Singularity for full pipeline reproducibility (please only use Conda as a last resort; see docs)
iii. Download the pipeline and test it on a minimal dataset with a single command
nextflow run nf-core/smartseq2 -profile test,<docker/singularity/conda/institute>Please check nf-core/configs to see if a custom config file to run nf-core pipelines already exists for your Institute. If so, you can simply use
-profile <institute>in your command. This will enable eitherdockerorsingularityand set the appropriate execution settings for your local compute environment.
iv. Start running your own analysis!
nextflow run nf-core/smartseq2 -profile <docker/singularity/conda/institute> --reads '*_R{1,2}.fastq.gz' --genome GRCh37See usage docs for all of the available options when running the pipeline.
Documentation
The nf-core/smartseq2 pipeline comes with documentation about the pipeline, found in the docs/ directory:
- Installation
- Pipeline configuration
- Running the pipeline
- Output and how to interpret the results
- Troubleshooting
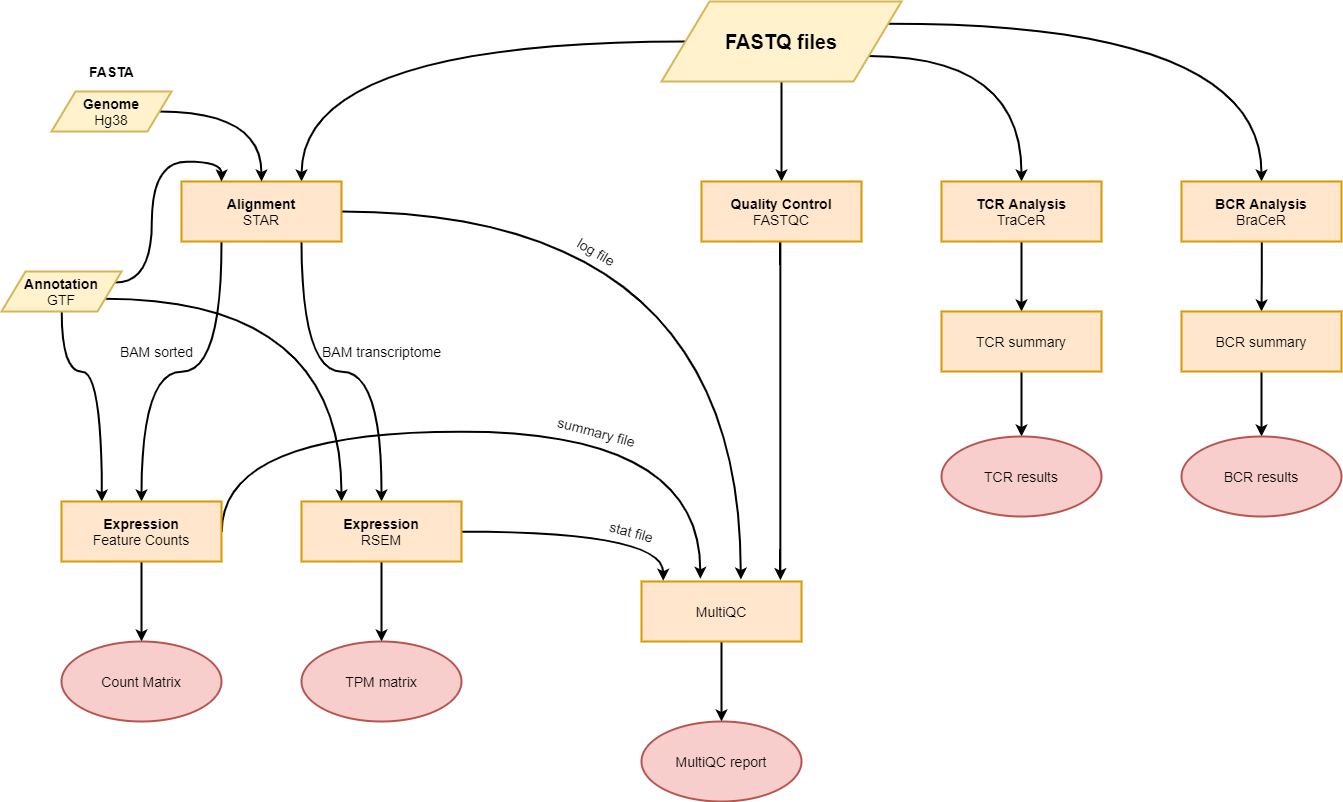
This pipeline was developed to process single cell RNA-seq data generated using the Smart-Seq2 protocol.
The pipeline features:
- Quality control using
fastqcandmultiqc - Alignment using
STAR - Quantification using
rsemorfeatureCounts - TCR analysis using
TraCeR. - BCR analysis using
BraCeR.
Credits
nf-core/smartseq2 was originally written by Sandro Carollo, Giorgos Fotakis and Gregor Sturm
Contributions and Support
If you would like to contribute to this pipeline, please see the contributing guidelines.
For further information or help, don’t hesitate to get in touch on Slack (you can join with this invite).
Citation
You can cite the nf-core publication as follows:
The nf-core framework for community-curated bioinformatics pipelines.
Philip Ewels, Alexander Peltzer, Sven Fillinger, Harshil Patel, Johannes Alneberg, Andreas Wilm, Maxime Ulysse Garcia, Paolo Di Tommaso & Sven Nahnsen.
Nat Biotechnol. 2020 Feb 13. doi: 10.1038/s41587-020-0439-x. ReadCube: Full Access Link