nf-core/stableexpression
This pipeline is dedicated to identifying the most stable genes within a single or multiple expression dataset(s). This is particularly useful for identifying the most suitable RT-qPCR reference genes for a specific species.
Introduction
nf-core/stableexpression is a bioinformatics pipeline aiming to aggregate multiple count datasets (public / provided by the user) for a specific species and find the most stable genes.
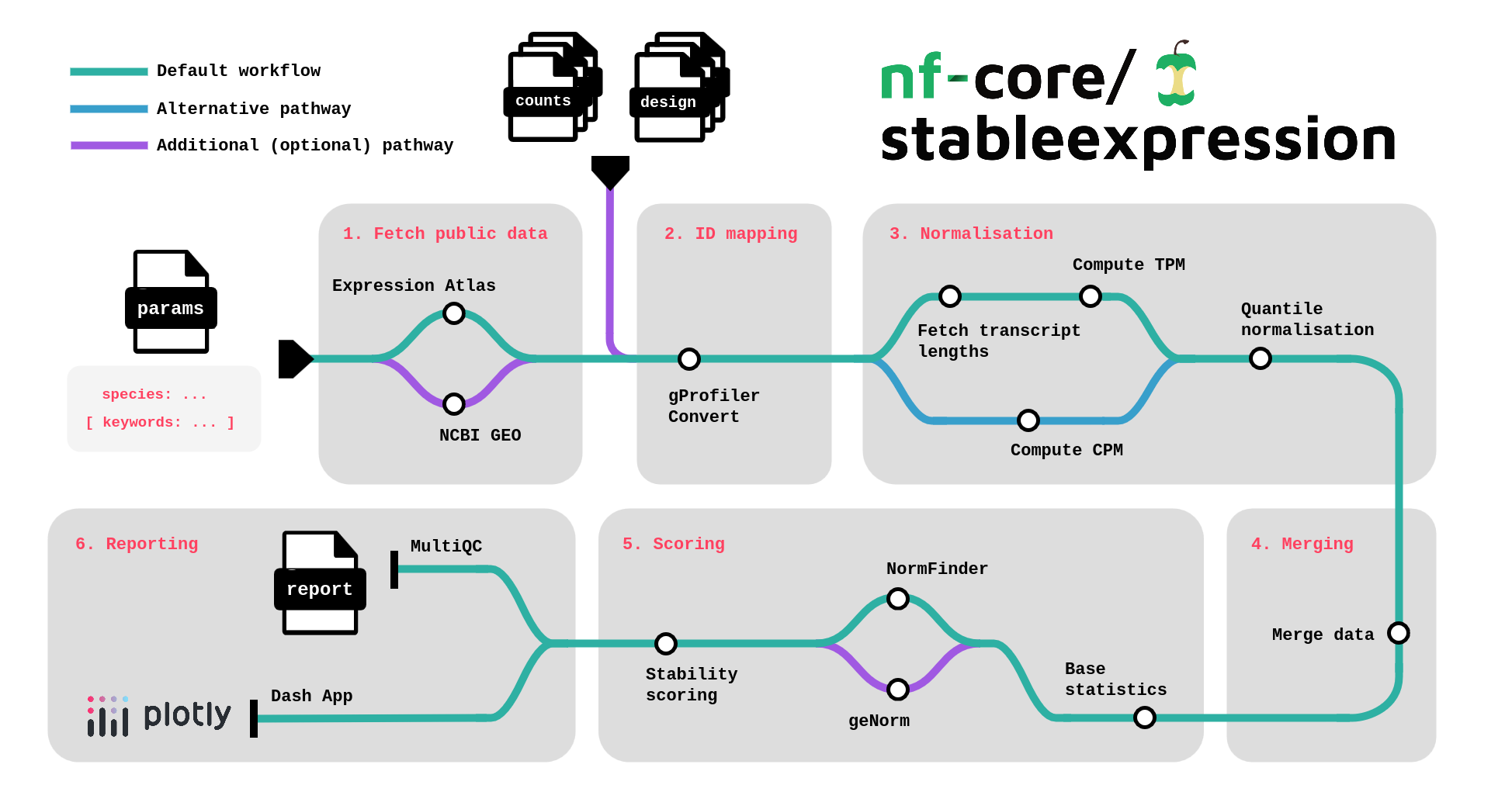
It takes as main inputs :
- a species name (mandatory)
- keywords for Expression Atlas / GEO search (optional)
- a CSV input file listing your own raw / normalised count datasets (optional).
Use cases:
- find the most suitable genes as RT-qPCR reference genes for a specific species (and optionally specific conditions)
- download all Expression Atlas and / or NCBI GEO datasets for a species (and optionally keywords)
Basic usage
If you are new to Nextflow and nf-core, please refer to this page on how to set-up Nextflow. Make sure to test your setup with -profile test before running the workflow on actual data.
To search the most stable genes in a species considering all public datasets, simply run:
nextflow run nf-core/stableexpression \
-profile <PROFILE (examples: docker / apptainer / conda / micromamba)> \
--species <SPECIES (examples: arabidopsis_thaliana / "drosophila melanogaster")> \
--outdir <OUTDIR (example: ./results)>More advanced usage
For more specific scenarios, like:
- fetching only specific conditions
- using your own expression dataset(s)
please refer to the usage documentation.
Profiles
See here for more information about profiles.
Pipeline output
To see the results of an example test run with a full size dataset refer to the results tab on the nf-core website pipeline page. For more details about the output files and reports, please refer to the output documentation.
Support us
If you like nf-core/stableexpression, please make sure you give it a star on GitHub.
Credits
nf-core/stableexpression was originally written by Olivier Coen.
Contributions and Support
If you would like to contribute to this pipeline, please see the contributing guidelines.
For further information or help, don’t hesitate to get in touch on the Slack #stableexpression channel (you can join with this invite).
Citations
An extensive list of references for the tools used by the pipeline can be found in the CITATIONS.md file.
You can cite the nf-core publication as follows:
The nf-core framework for community-curated bioinformatics pipelines.
Philip Ewels, Alexander Peltzer, Sven Fillinger, Harshil Patel, Johannes Alneberg, Andreas Wilm, Maxime Ulysse Garcia, Paolo Di Tommaso & Sven Nahnsen.
Nat Biotechnol. 2020 Feb 13. doi: 10.1038/s41587-020-0439-x.
