nf-core/viralrecon
Assembly and intrahost/low-frequency variant calling for viral samples
2.6.0). The latest
stable release is
3.0.0
.
Introduction
nf-core/viralrecon is a bioinformatics analysis pipeline used to perform assembly and intra-host/low-frequency variant calling for viral samples. The pipeline supports both Illumina and Nanopore sequencing data. For Illumina short-reads the pipeline is able to analyse metagenomics data typically obtained from shotgun sequencing (e.g. directly from clinical samples) and enrichment-based library preparation methods (e.g. amplicon-based: ARTIC SARS-CoV-2 enrichment protocol; or probe-capture-based). For Nanopore data the pipeline only supports amplicon-based analysis obtained from primer sets created and maintained by the ARTIC Network.
On release, automated continuous integration tests run the pipeline on a full-sized dataset on the AWS cloud infrastructure. This ensures that the pipeline runs on AWS, has sensible resource allocation defaults set to run on real-world datasets, and permits the persistent storage of results to benchmark between pipeline releases and other analysis sources. The results obtained from running the full-sized tests individually for each --platform option can be viewed on the nf-core website and the output directories will be named accordingly i.e. platform_illumina/ and platform_nanopore/.
The pipeline is built using Nextflow, a workflow tool to run tasks across multiple compute infrastructures in a very portable manner. It uses Docker/Singularity containers making installation trivial and results highly reproducible. The Nextflow DSL2 implementation of this pipeline uses one container per process which makes it much easier to maintain and update software dependencies. Where possible, these processes have been submitted to and installed from nf-core/modules in order to make them available to all nf-core pipelines, and to everyone within the Nextflow community!
Pipeline summary
The pipeline has numerous options to allow you to run only specific aspects of the workflow if you so wish. For example, for Illumina data you can skip the host read filtering step with Kraken 2 with --skip_kraken2 or you can skip all of the assembly steps with the --skip_assembly parameter. See the usage and parameter docs for all of the available options when running the pipeline.
The SRA download functionality has been removed from the pipeline (>=2.1) and ported to an independent workflow called nf-core/fetchngs. You can provide --nf_core_pipeline viralrecon when running nf-core/fetchngs to download and auto-create a samplesheet containing publicly available samples that can be accepted directly by the Illumina processing mode of nf-core/viralrecon.
A number of improvements were made to the pipeline recently, mainly with regard to the variant calling. Please see Major updates in v2.3 for a more detailed description.
Illumina
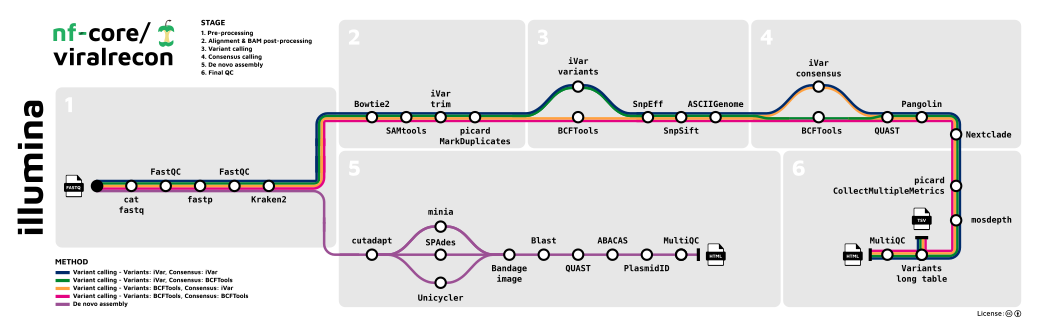
- Merge re-sequenced FastQ files (
cat) - Read QC (
FastQC) - Adapter trimming (
fastp) - Removal of host reads (
Kraken 2; optional) - Variant calling
- Read alignment (
Bowtie 2) - Sort and index alignments (
SAMtools) - Primer sequence removal (
iVar; amplicon data only) - Duplicate read marking (
picard; optional) - Alignment-level QC (
picard,SAMtools) - Genome-wide and amplicon coverage QC plots (
mosdepth) - Choice of multiple variant callers (
iVar variants; default for amplicon data ||BCFTools; default for metagenomics data)- Variant annotation (
SnpEff,SnpSift) - Individual variant screenshots with annotation tracks (
ASCIIGenome)
- Variant annotation (
- Choice of multiple consensus callers (
BCFTools,BEDTools; default for both amplicon and metagenomics data ||iVar consensus) - Create variants long format table collating per-sample information for individual variants (
BCFTools), functional effect prediction (SnpSift) and lineage analysis (Pangolin)
- Read alignment (
- De novo assembly
- Present QC and visualisation for raw read, alignment, assembly and variant calling results (
MultiQC)
Nanopore
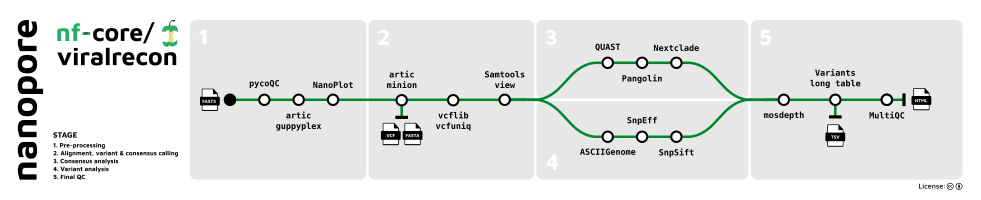
- Sequencing QC (
pycoQC) - Aggregate pre-demultiplexed reads from MinKNOW/Guppy (
artic guppyplex) - Read QC (
NanoPlot) - Align reads, call variants and generate consensus sequence (
artic minion) - Remove unmapped reads and obtain alignment metrics (
SAMtools) - Genome-wide and amplicon coverage QC plots (
mosdepth) - Downstream variant analysis:
- Count metrics (
BCFTools) - Variant annotation (
SnpEff,SnpSift) - Consensus assessment report (
QUAST) - Lineage analysis (
Pangolin) - Clade assignment, mutation calling and sequence quality checks (
Nextclade) - Individual variant screenshots with annotation tracks (
ASCIIGenome) - Create variants long format table collating per-sample information for individual variants (
BCFTools), functional effect prediction (SnpSift) and lineage analysis (Pangolin)
- Count metrics (
- Present QC, visualisation and custom reporting for sequencing, raw reads, alignment and variant calling results (
MultiQC)
Quick Start
-
Install
Nextflow(>=22.10.1) -
Install any of
Docker,Singularity(you can follow this tutorial),Podman,ShifterorCharliecloudfor full pipeline reproducibility (you can useCondaboth to install Nextflow itself and also to manage software within pipelines. Please only use it within pipelines as a last resort; see docs). -
Download the pipeline and test it on a minimal dataset with a single command:
nextflow run nf-core/viralrecon -profile test,YOURPROFILE --outdir <OUTDIR>Note that some form of configuration will be needed so that Nextflow knows how to fetch the required software. This is usually done in the form of a config profile (
YOURPROFILEin the example command above). You can chain multiple config profiles in a comma-separated string.- The pipeline comes with config profiles called
docker,singularity,podman,shifter,charliecloudandcondawhich instruct the pipeline to use the named tool for software management. For example,-profile test,docker. - Please check nf-core/configs to see if a custom config file to run nf-core pipelines already exists for your Institute. If so, you can simply use
-profile <institute>in your command. This will enable eitherdockerorsingularityand set the appropriate execution settings for your local compute environment. - If you are using
singularity, please use thenf-core downloadcommand to download images first, before running the pipeline. Setting theNXF_SINGULARITY_CACHEDIRorsingularity.cacheDirNextflow options enables you to store and re-use the images from a central location for future pipeline runs. - If you are using
conda, it is highly recommended to use theNXF_CONDA_CACHEDIRorconda.cacheDirsettings to store the environments in a central location for future pipeline runs.
- The pipeline comes with config profiles called
-
Start running your own analysis!
- Please provide pipeline parameters via the CLI or Nextflow
-params-fileoption. Custom config files including those provided by the-cNextflow option can be used to provide any configuration except for parameters; see docs.
-
Typical command for Illumina shotgun analysis:
nextflow run nf-core/viralrecon \ --input samplesheet.csv \ --outdir <OUTDIR> \ --platform illumina \ --protocol metagenomic \ --genome 'MN908947.3' \ -profile <docker/singularity/podman/conda/institute> -
Typical command for Illumina amplicon analysis:
nextflow run nf-core/viralrecon \ --input samplesheet.csv \ --outdir <OUTDIR> \ --platform illumina \ --protocol amplicon \ --genome 'MN908947.3' \ --primer_set artic \ --primer_set_version 3 \ --skip_assembly \ -profile <docker/singularity/podman/conda/institute> -
Typical command for Nanopore amplicon analysis:
nextflow run nf-core/viralrecon \ --input samplesheet.csv \ --outdir <OUTDIR> \ --platform nanopore \ --genome 'MN908947.3' \ --primer_set_version 3 \ --fastq_dir fastq_pass/ \ --fast5_dir fast5_pass/ \ --sequencing_summary sequencing_summary.txt \ -profile <docker/singularity/podman/conda/institute> -
An executable Python script called
fastq_dir_to_samplesheet.pyhas been provided if you are using--platform illuminaand would like to auto-create an input samplesheet based on a directory containing FastQ files before you run the pipeline (requires Python 3 installed locally) e.g.wget -L https://raw.githubusercontent.com/nf-core/viralrecon/master/bin/fastq_dir_to_samplesheet.py ./fastq_dir_to_samplesheet.py <FASTQ_DIR> samplesheet.csv -
You can find the default keys used to specify
--genomein the genomes config file. This provides default options for-
Reference genomes (including SARS-CoV-2)
-
Genome associates primer sets
-
The Pangolin and Nextclade lineage and clade definitions change regularly as new SARS-CoV-2 lineages are discovered. For instructions to use more recent versions of lineage analysis tools like Pangolin and Nextclade please refer to the updating containers section in the usage docs.
Where possible we are trying to collate links and settings for standard primer sets to make it easier to run the pipeline with standard keys; see usage docs.
-
- Please provide pipeline parameters via the CLI or Nextflow
Documentation
The nf-core/viralrecon pipeline comes with documentation about the pipeline usage, parameters and output.
Credits
These scripts were originally written by Sarai Varona, Miguel Juliá, Erika Kvalem and Sara Monzon from BU-ISCIII and co-ordinated by Isabel Cuesta for the Institute of Health Carlos III, Spain. Through collaboration with the nf-core community the pipeline has now been updated substantially to include additional processing steps, to standardise inputs/outputs and to improve pipeline reporting; implemented and maintained primarily by Harshil Patel (@drpatelh) from Seqera Labs, Spain.
The key steps in the Nanopore implementation of the pipeline are carried out using the ARTIC Network’s field bioinformatics pipeline and were inspired by the amazing work carried out by contributors to the connor-lab/ncov2019-artic-nf pipeline originally written by Matt Bull for use by the COG-UK project. Thank you for all of your incredible efforts during this pandemic!
Many thanks to others who have helped out and contributed along the way too, including (but not limited to)*:
* Listed in alphabetical order
Contributions and Support
If you would like to contribute to this pipeline, please see the contributing guidelines.
For further information or help, don’t hesitate to get in touch on the Slack #viralrecon channel (you can join with this invite).
Citations
If you use nf-core/viralrecon for your analysis, please cite it using the following doi: 10.5281/zenodo.3901628
An extensive list of references for the tools used by the pipeline can be found in the CITATIONS.md file.
You can cite the nf-core publication as follows:
The nf-core framework for community-curated bioinformatics pipelines.
Philip Ewels, Alexander Peltzer, Sven Fillinger, Harshil Patel, Johannes Alneberg, Andreas Wilm, Maxime Ulysse Garcia, Paolo Di Tommaso & Sven Nahnsen.
Nat Biotechnol. 2020 Feb 13. doi: 10.1038/s41587-020-0439-x.