nf-core/ampliseq
Amplicon sequencing analysis workflow using DADA2 and QIIME2
2.1.0). The latest
stable release is
2.16.1
.
Usage
Input specifications
The input data can be passed to nf-core/ampliseq in three possible ways using the --input parameter, either a folder containing zipped FastQ files, a tab-seperated samplesheet, or a fasta file to be taxonomically classified.
Optionally, a metadata sheet can be specified for downstream analysis.
Direct FASTQ input
The easiest way is to specify directly the path to the folder that contains your input FASTQ files. For example:
--input 'path/to/data/'File names must follow a specific pattern, default is /*_R{1,2}_001.fastq.gz, but this can be adjusted with --extension.
For example, the following files in folder data would be processed as sample1 and sample2:
data
|-sample1_1_L001_R1_001.fastq.gz
|-sample1_1_L001_R2_001.fastq.gz
|-sample2_1_L001_R1_001.fastq.gz
|-sample2_1_L001_R2_001.fastq.gzAll sequencing data should originate from one sequencing run, because processing relies on run-specific error models that are unreliable when data from several sequencing runs are mixed. Sequencing data originating from multiple sequencing runs requires additionally the parameter --multiple_sequencing_runs and a specific folder structure, for example:
data
|-runA
| |-sample1_1_L001_R1_001.fastq.gz
| |-sample1_1_L001_R2_001.fastq.gz
| |-sample2_1_L001_R1_001.fastq.gz
| |-sample2_1_L001_R2_001.fastq.gz
|
|-runB
|-sample3_1_L001_R1_001.fastq.gz
|-sample3_1_L001_R2_001.fastq.gz
|-sample4_1_L001_R1_001.fastq.gz
|-sample4_1_L001_R2_001.fastq.gzWhere sample1 and sample2 were sequenced in one sequencing run and sample3 and sample4 in another sequencing run.
Please note the following additional requirements:
- Files names must be unique
- Valid file extensions:
.fastq.gz,.fq.gz(files must be compressed) - The path must be enclosed in quotes
--extensionmust have at least one*wildcard character- When using the pipeline with paired end data, the
--extensionmust use{1,2}(or similar) notation to specify read pairs - To run single-end data you must additionally specify
--single_end - Sample identifiers are extracted from file names, i.e. the string before the first underscore
_, these must be unique (also across sequencing runs) - If your data is scattered, produce a sample sheet
Samplesheet input
The sample sheet file is an alternative way to provide input reads, it must be a tab-separated file ending with .tsv that must have two to four columns with the following headers:
| Column | Necessity | Description |
|---|---|---|
| sampleID | required | Unique sample identifiers, any unique string (may not contain dots .) |
| forwardReads | required | Paths to (forward) reads zipped FastQ files |
| reverseReads | optional | Paths to reverse reads zipped FastQ files, required if the data is paired-end |
| run | optional | If the data was produced by multiple sequencing runs, any string |
--input 'path/to/samplesheet.tsv'For example, the samplesheet may contain:
sampleID forwardReads reverseReads run
sample1 ./data/S1_R1_001.fastq.gz ./data/S1_R2_001.fastq.gz A
sample2 ./data/S2_fw.fastq.gz ./data/S2_rv.fastq.gz A
sample3 ./S4x.fastq.gz ./S4y.fastq.gz B
sample4 ./a.fastq.gz ./b.fastq.gz BPlease note the following requirements:
- 2 to 4 tab-seperated columns
- Valid file extension:
.tsv - Must contain the header
sampleIDandforwardReads - May contain the header
reverseReadsandrun - Sample IDs must be unique
- FastQ files must be compressed (
.fastq.gz,.fq.gz) - Within one samplesheet, only one type of raw data should be specified (same amplicon & sequencing method)
An example samplesheet has been provided with the pipeline.
Fasta file input, to taxonomically classify previously produced ASV/OTU sequences
When pointing at a file ending with .fasta, .fna or .fa, the containing sequences will be taxonomically classified. All other pipeline steps will be skipped.
--input 'path/to/amplicon_sequences.fasta'Please note the following requirements:
- Valid file extensions:
.fasta,.fnaor.fa
Metadata
The metadata file must be tab-separated with a header line, for more details please see the nf-core/ampliseq website documentation.
--metadata "path/to/metadata.tsv"For example:
ID condition
sample1 control
sample2 treatment
sample3 control
sample4 treatmentRunning the pipeline
The typical command for running the pipeline is as follows:
nextflow run nf-core/ampliseq \
-profile singularity \
--input "data" \
--FW_primer GTGYCAGCMGCCGCGGTAA \
--RV_primer GGACTACNVGGGTWTCTAAT \
--metadata "data/Metadata.tsv"This will launch the pipeline with the singularity configuration profile. See below -profile for more information about profiles.
Note that the pipeline will create the following files in your working directory:
work # Directory containing the nextflow working files
results # Finished results (configurable, see below)
.nextflow_log # Log file from Nextflow
# Other nextflow hidden files, eg. history of pipeline runs and old logs.NB: If the data originates from multiple sequencing runs, the error profile of each of those sequencing runs needs to be considered separately. Using the
runcolumn in the samplesheet input or adding--multiple_sequencing_runsfor Direct FASTQ input will separate certain processes by the sequencing run. Please see the following example:
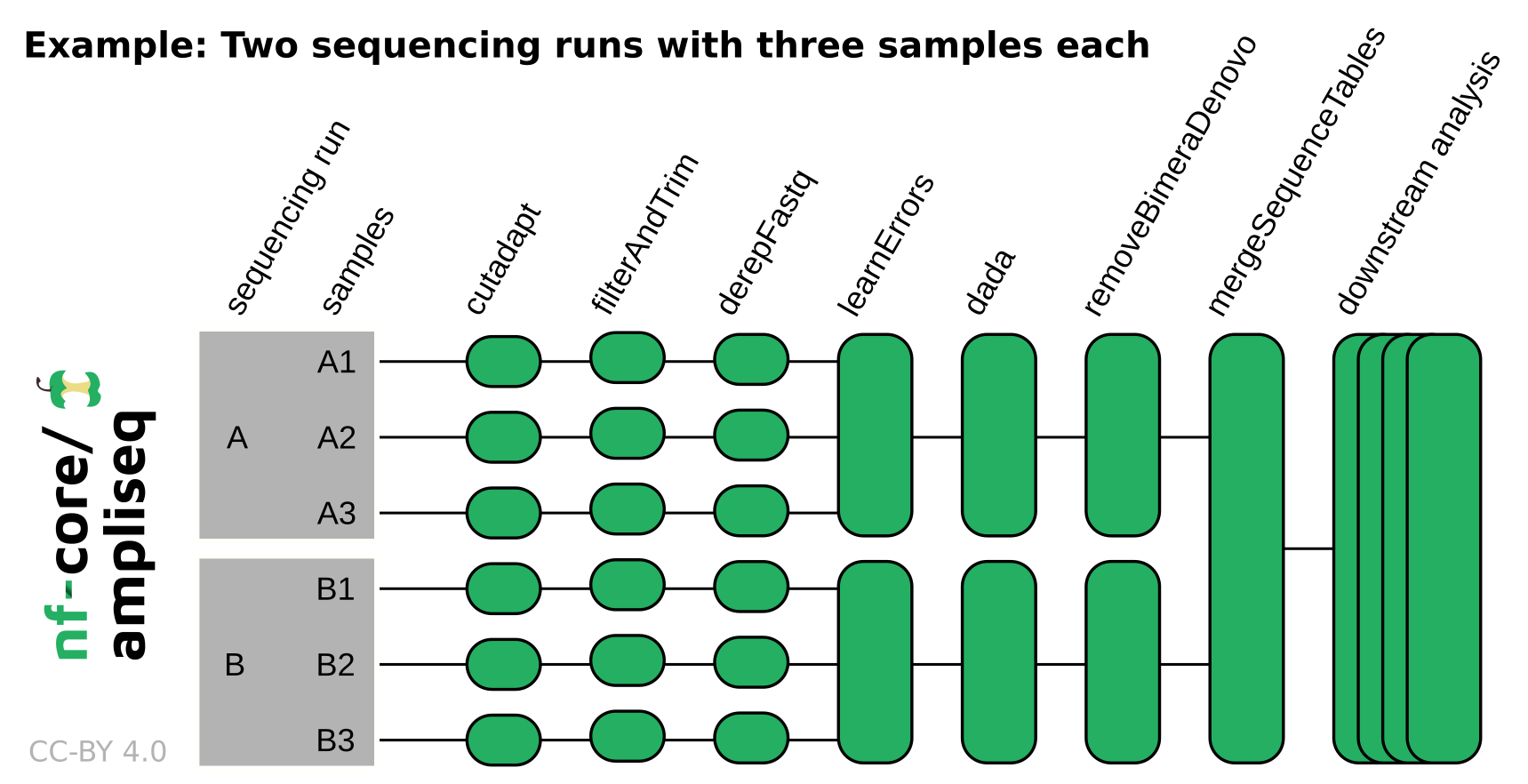
See the nf-core/ampliseq website documentation for more information about pipeline specific parameters.
Updating the pipeline
When you run the above command, Nextflow automatically pulls the pipeline code from GitHub and stores it as a cached version. When running the pipeline after this, it will always use the cached version if available - even if the pipeline has been updated since. To make sure that you’re running the latest version of the pipeline, make sure that you regularly update the cached version of the pipeline:
nextflow pull nf-core/ampliseqReproducibility
It is a good idea to specify a pipeline version when running the pipeline on your data. This ensures that a specific version of the pipeline code and software are used when you run your pipeline. If you keep using the same tag, you’ll be running the same version of the pipeline, even if there have been changes to the code since.
First, go to the nf-core/ampliseq releases page and find the latest version number - numeric only (eg. 2.0.0). Then specify this when running the pipeline with -r (one hyphen) - eg. -r 2.0.0.
This version number will be logged in reports when you run the pipeline, so that you’ll know what you used when you look back in the future.
Core Nextflow arguments
NB: These options are part of Nextflow and use a single hyphen (pipeline parameters use a double-hyphen).
-profile
Use this parameter to choose a configuration profile. Profiles can give configuration presets for different compute environments.
Several generic profiles are bundled with the pipeline which instruct the pipeline to use software packaged using different methods (Docker, Singularity, Podman, Shifter, Charliecloud, Conda) - see below. When using Biocontainers, most of these software packaging methods pull Docker containers from quay.io e.g FastQC except for Singularity which directly downloads Singularity images via https hosted by the Galaxy project and Conda which downloads and installs software locally from Bioconda.
We highly recommend the use of Docker or Singularity containers for full pipeline reproducibility, however when this is not possible, Conda is also supported.
The pipeline also dynamically loads configurations from https://github.com/nf-core/configs when it runs, making multiple config profiles for various institutional clusters available at run time. For more information and to see if your system is available in these configs please see the nf-core/configs documentation.
Note that multiple profiles can be loaded, for example: -profile test,docker - the order of arguments is important!
They are loaded in sequence, so later profiles can overwrite earlier profiles.
If -profile is not specified, the pipeline will run locally and expect all software to be installed and available on the PATH. This is not recommended.
docker- A generic configuration profile to be used with Docker
singularity- A generic configuration profile to be used with Singularity
podman- A generic configuration profile to be used with Podman
shifter- A generic configuration profile to be used with Shifter
charliecloud- A generic configuration profile to be used with Charliecloud
conda- A generic configuration profile to be used with Conda. Please only use Conda as a last resort i.e. when it’s not possible to run the pipeline with Docker, Singularity, Podman, Shifter or Charliecloud.
test,test_multi,test_full,test_pacbio_its,test_iontorrent,test_doubleprimers- Profiles with a complete configuration for automated testing
- Includes links to test data so needs no other parameters
-resume
Specify this when restarting a pipeline. Nextflow will used cached results from any pipeline steps where the inputs are the same, continuing from where it got to previously.
You can also supply a run name to resume a specific run: -resume [run-name]. Use the nextflow log command to show previous run names.
-c
Specify the path to a specific config file (this is a core Nextflow command). See the nf-core website documentation for more information.
Custom configuration
Resource requests
Whilst the default requirements set within the pipeline will hopefully work for most people and with most input data, you may find that you want to customise the compute resources that the pipeline requests. Each step in the pipeline has a default set of requirements for number of CPUs, memory and time. For most of the steps in the pipeline, if the job exits with any of the error codes specified here it will automatically be resubmitted with higher requests (2 x original, then 3 x original). If it still fails after the third attempt then the pipeline execution is stopped.
For example, if the nf-core/rnaseq pipeline is failing after multiple re-submissions of the STAR_ALIGN process due to an exit code of 137 this would indicate that there is an out of memory issue:
[62/149eb0] NOTE: Process `RNASEQ:ALIGN_STAR:STAR_ALIGN (WT_REP1)` terminated with an error exit status (137) -- Execution is retried (1)
Error executing process > 'RNASEQ:ALIGN_STAR:STAR_ALIGN (WT_REP1)'
Caused by:
Process `RNASEQ:ALIGN_STAR:STAR_ALIGN (WT_REP1)` terminated with an error exit status (137)
Command executed:
STAR \
--genomeDir star \
--readFilesIn WT_REP1_trimmed.fq.gz \
--runThreadN 2 \
--outFileNamePrefix WT_REP1. \
<TRUNCATED>
Command exit status:
137
Command output:
(empty)
Command error:
.command.sh: line 9: 30 Killed STAR --genomeDir star --readFilesIn WT_REP1_trimmed.fq.gz --runThreadN 2 --outFileNamePrefix WT_REP1. <TRUNCATED>
Work dir:
/home/pipelinetest/work/9d/172ca5881234073e8d76f2a19c88fb
Tip: you can replicate the issue by changing to the process work dir and entering the command `bash .command.run`To bypass this error you would need to find exactly which resources are set by the STAR_ALIGN process. The quickest way is to search for process STAR_ALIGN in the nf-core/rnaseq Github repo. We have standardised the structure of Nextflow DSL2 pipelines such that all module files will be present in the modules/ directory and so based on the search results the file we want is modules/nf-core/software/star/align/main.nf. If you click on the link to that file you will notice that there is a label directive at the top of the module that is set to label process_high. The Nextflow label directive allows us to organise workflow processes in separate groups which can be referenced in a configuration file to select and configure subset of processes having similar computing requirements. The default values for the process_high label are set in the pipeline’s base.config which in this case is defined as 72GB. Providing you haven’t set any other standard nf-core parameters to cap the maximum resources used by the pipeline then we can try and bypass the STAR_ALIGN process failure by creating a custom config file that sets at least 72GB of memory, in this case increased to 100GB. The custom config below can then be provided to the pipeline via the -c parameter as highlighted in previous sections.
process {
withName: STAR_ALIGN {
memory = 100.GB
}
}NB: We specify just the process name i.e.
STAR_ALIGNin the config file and not the full task name string that is printed to screen in the error message or on the terminal whilst the pipeline is running i.e.RNASEQ:ALIGN_STAR:STAR_ALIGN. You may get a warning suggesting that the process selector isn’t recognised but you can ignore that if the process name has been specified correctly. This is something that needs to be fixed upstream in core Nextflow.
Tool-specific options
For the ultimate flexibility, we have implemented and are using Nextflow DSL2 modules in a way where it is possible for both developers and users to change tool-specific command-line arguments (e.g. providing an additional command-line argument to the STAR_ALIGN process) as well as publishing options (e.g. saving files produced by the STAR_ALIGN process that aren’t saved by default by the pipeline). In the majority of instances, as a user you won’t have to change the default options set by the pipeline developer(s), however, there may be edge cases where creating a simple custom config file can improve the behaviour of the pipeline if for example it is failing due to a weird error that requires setting a tool-specific parameter to deal with smaller / larger genomes.
The command-line arguments passed to STAR in the STAR_ALIGN module are a combination of:
-
Mandatory arguments or those that need to be evaluated within the scope of the module, as supplied in the
scriptsection of the module file. -
An
options.argsstring of non-mandatory parameters that is set to be empty by default in the module but can be overwritten when including the module in the sub-workflow / workflow context via theaddParamsNextflow option.
The nf-core/rnaseq pipeline has a sub-workflow (see terminology) specifically to align reads with STAR and to sort, index and generate some basic stats on the resulting BAM files using SAMtools. At the top of this file we import the STAR_ALIGN module via the Nextflow include keyword and by default the options passed to the module via the addParams option are set as an empty Groovy map here; this in turn means options.args will be set to empty by default in the module file too. This is an intentional design choice and allows us to implement well-written sub-workflows composed of a chain of tools that by default run with the bare minimum parameter set for any given tool in order to make it much easier to share across pipelines and to provide the flexibility for users and developers to customise any non-mandatory arguments.
When including the sub-workflow above in the main pipeline workflow we use the same include statement, however, we now have the ability to overwrite options for each of the tools in the sub-workflow including the align_options variable that will be used specifically to overwrite the optional arguments passed to the STAR_ALIGN module. In this case, the options to be provided to STAR_ALIGN have been assigned sensible defaults by the developer(s) in the pipeline’s modules.config and can be accessed and customised in the workflow context too before eventually passing them to the sub-workflow as a Groovy map called star_align_options. These options will then be propagated from workflow -> sub-workflow -> module.
As mentioned at the beginning of this section it may also be necessary for users to overwrite the options passed to modules to be able to customise specific aspects of the way in which a particular tool is executed by the pipeline. Given that all of the default module options are stored in the pipeline’s modules.config as a params variable it is also possible to overwrite any of these options via a custom config file.
Say for example we want to append an additional, non-mandatory parameter (i.e. --outFilterMismatchNmax 16) to the arguments passed to the STAR_ALIGN module. Firstly, we need to copy across the default args specified in the modules.config and create a custom config file that is a composite of the default args as well as the additional options you would like to provide. This is very important because Nextflow will overwrite the default value of args that you provide via the custom config.
As you will see in the example below, we have:
- appended
--outFilterMismatchNmax 16to the defaultargsused by the module. - changed the default
publish_dirvalue to where the files will eventually be published in the main results directory. - appended
'bam':''to the default value ofpublish_filesso that the BAM files generated by the process will also be saved in the top-level results directory for the module. Note:'out':'log'means any file/directory ending inoutwill now be saved in a separate directory calledmy_star_directory/log/.
params {
modules {
'star_align' {
args = "--quantMode TranscriptomeSAM --twopassMode Basic --outSAMtype BAM Unsorted --readFilesCommand zcat --runRNGseed 0 --outFilterMultimapNmax 20 --alignSJDBoverhangMin 1 --outSAMattributes NH HI AS NM MD --quantTranscriptomeBan Singleend --outFilterMismatchNmax 16"
publish_dir = "my_star_directory"
publish_files = ['out':'log', 'tab':'log', 'bam':'']
}
}
}Updating containers
The Nextflow DSL2 implementation of this pipeline uses one container per process which makes it much easier to maintain and update software dependencies. If for some reason you need to use a different version of a particular tool with the pipeline then you just need to identify the process name and override the Nextflow container definition for that process using the withName declaration. For example, in the nf-core/viralrecon pipeline a tool called Pangolin has been used during the COVID-19 pandemic to assign lineages to SARS-CoV-2 genome sequenced samples. Given that the lineage assignments change quite frequently it doesn’t make sense to re-release the nf-core/viralrecon everytime a new version of Pangolin has been released. However, you can override the default container used by the pipeline by creating a custom config file and passing it as a command-line argument via -c custom.config.
-
Check the default version used by the pipeline in the module file for Pangolin
-
Find the latest version of the Biocontainer available on Quay.io
-
Create the custom config accordingly:
-
For Docker:
process { withName: PANGOLIN { container = 'quay.io/biocontainers/pangolin:3.0.5--pyhdfd78af_0' } } -
For Singularity:
process { withName: PANGOLIN { container = 'https://depot.galaxyproject.org/singularity/pangolin:3.0.5--pyhdfd78af_0' } } -
For Conda:
process { withName: PANGOLIN { conda = 'bioconda::pangolin=3.0.5' } }
-
NB: If you wish to periodically update individual tool-specific results (e.g. Pangolin) generated by the pipeline then you must ensure to keep the
work/directory otherwise the-resumeability of the pipeline will be compromised and it will restart from scratch.
nf-core/configs
In most cases, you will only need to create a custom config as a one-off but if you and others within your organisation are likely to be running nf-core pipelines regularly and need to use the same settings regularly it may be a good idea to request that your custom config file is uploaded to the nf-core/configs git repository. Before you do this please can you test that the config file works with your pipeline of choice using the -c parameter. You can then create a pull request to the nf-core/configs repository with the addition of your config file, associated documentation file (see examples in nf-core/configs/docs), and amending nfcore_custom.config to include your custom profile.
See the main Nextflow documentation for more information about creating your own configuration files.
If you have any questions or issues please send us a message on Slack on the #configs channel.
Running in the background
Nextflow handles job submissions and supervises the running jobs. The Nextflow process must run until the pipeline is finished.
The Nextflow -bg flag launches Nextflow in the background, detached from your terminal so that the workflow does not stop if you log out of your session. The logs are saved to a file.
Alternatively, you can use screen / tmux or similar tool to create a detached session which you can log back into at a later time.
Some HPC setups also allow you to run nextflow within a cluster job submitted your job scheduler (from where it submits more jobs).
Nextflow memory requirements
In some cases, the Nextflow Java virtual machines can start to request a large amount of memory.
We recommend adding the following line to your environment to limit this (typically in ~/.bashrc or ~./bash_profile):
NXF_OPTS='-Xms1g -Xmx4g'