nf-core/bacass
Simple bacterial assembly and annotation pipeline
1.1.0). The latest
stable release is
2.5.0
.
Output
This document describes the output produced by the pipeline. Most of the plots are taken from the MultiQC report, which summarises results at the end of the pipeline.
Pipeline overview
The pipeline is built using Nextflow and processes data using the following steps:
Quality trimming and QC
Short Read Trimming
This step quality trims the end of reads, removes degenerate or too short reads and if needed, combines reads coming from multiple sequencing runs.
Output directory: {sample_id}/trimming/shortreads/
*.fastq.gz- trimmed (and combined reads)
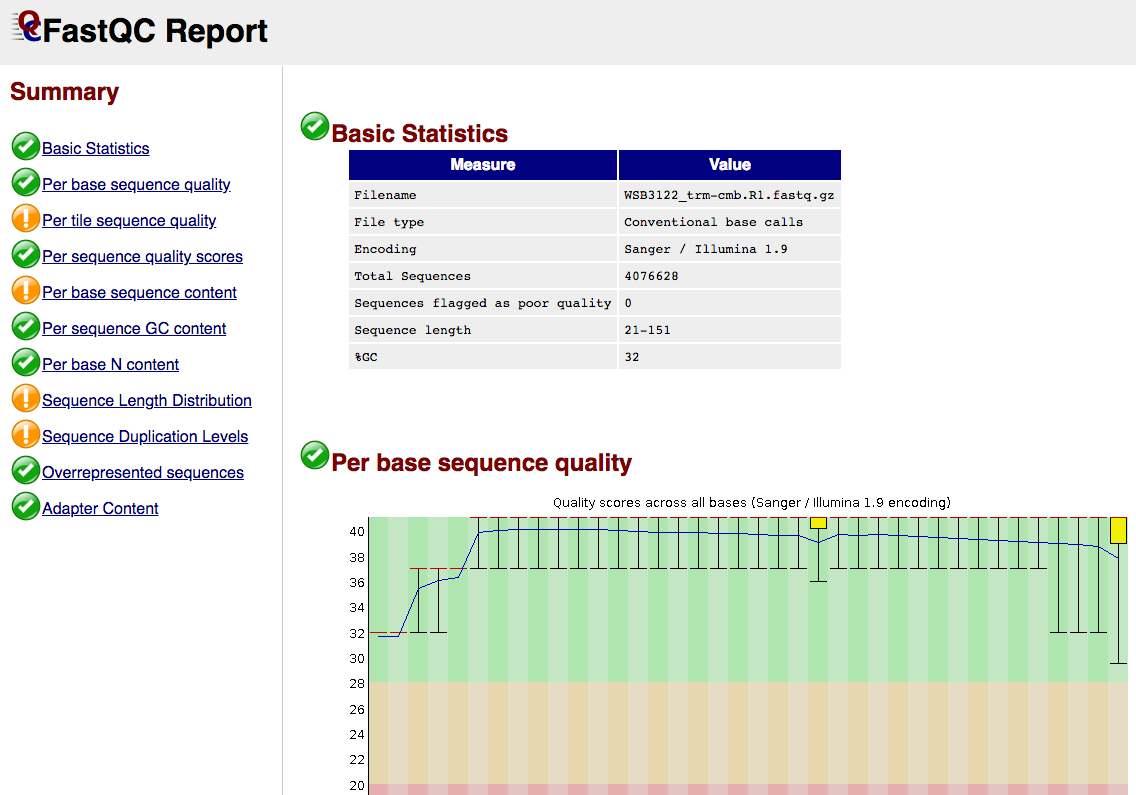
Short Read RAW QC
This step runs FastQC which produces general quality metrics on your (trimmed) samples and plots them.
Output directory: {sample_id}/trimming/shortreads/
*_fastqc.html- FastQC report, containing quality metrics for your trimmed reads
*_fastqc.zip- zip file containing the FastQC report, tab-delimited data file and plot images
For further reading and documentation see the FastQC help.
Long Read Trimming
This step performs long read trimming on Nanopore input (if provided).
Output directory: {sample_id}/trimming/longreads/
trimmed.fastq- The trimmed FASTQ file
Long Read RAW QC
These steps perform long read QC for input data (if provided).
Output directory: {sample_id}/QC_Longreads/
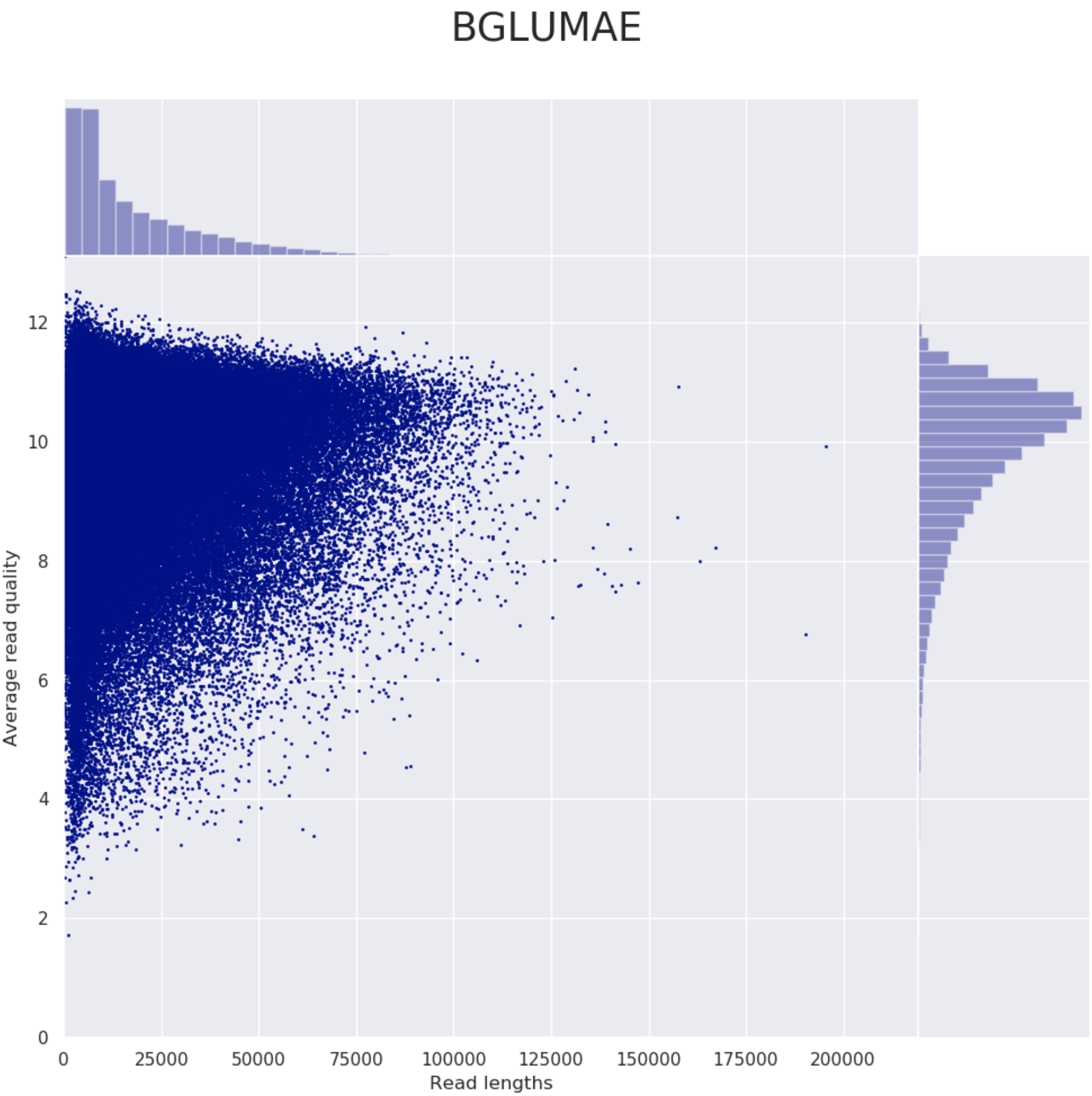
NanoPlotPycoQC
Please refer to the documentation of NanoPlot and PycoQC if you want to know more about the plots created by these tools.
Example plot from Nanoplot:
Taxonomic classification
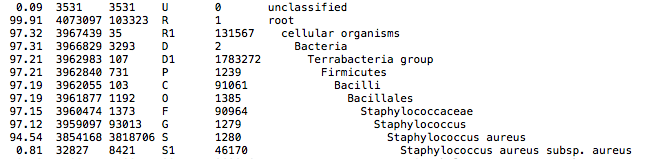
This QC step classifies your reads using Kraken2 a k-mer based approach. This helps to identify samples that have purity issues. Ideally you will not want to assemble reads from samples that are contaminated or contain multiple species. If you like to visualize the report, try Pavian or Krakey.
Output directory: {sample}/
*_kraken2.report- Classification in the Kraken(1) report format. See webpage for more details
Kraken2 report screenshot
Assembly Output
Trimmed reads are assembled with Unicycler in short or hybrid assembly modes. For long-read assembly, there are also canu and miniasm available.
Unicycler is a pipeline on its own, which at least for Illumina reads mainly acts as a frontend to Spades with added polishing steps.
Output directory: {sample_id}/unicycler
{sample}_assembly.fasta- Final assembly
{sample}_assembly.gfa- Final assembly in Graphical Fragment Assembly (GFA) format
{sample}_unicycler.log- Log file summarizing steps and intermediate results on the Unicycler execution
Check out the Unicycler documentation for more information on Unicycler output.
Output directory: {sample_id}/canu
Check out the Canu documentation for more information on Canu output.
Output directory: {sample_id}/miniasm
consensus- The consensus sequence created by
miniasm
- The consensus sequence created by
Check out the Miniasm documentation for more information on Miniasm output.
Assembly Visualization with Bandage
The GFA file produced in the assembly step with Unicycler can be used to visualise the assembly graph, which is done here with Bandage. We highly recommend to run the Bandage GUI for more versatile visualisation options (annotations etc).
Output directory: {sample_id}/unicycler
{sample}_assembly.png- Bandage visualization of assembly

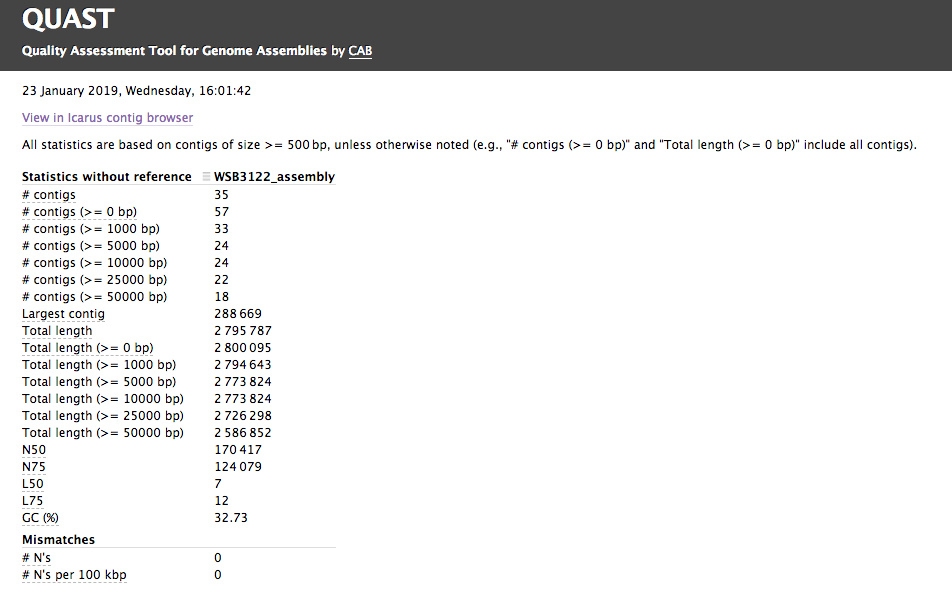
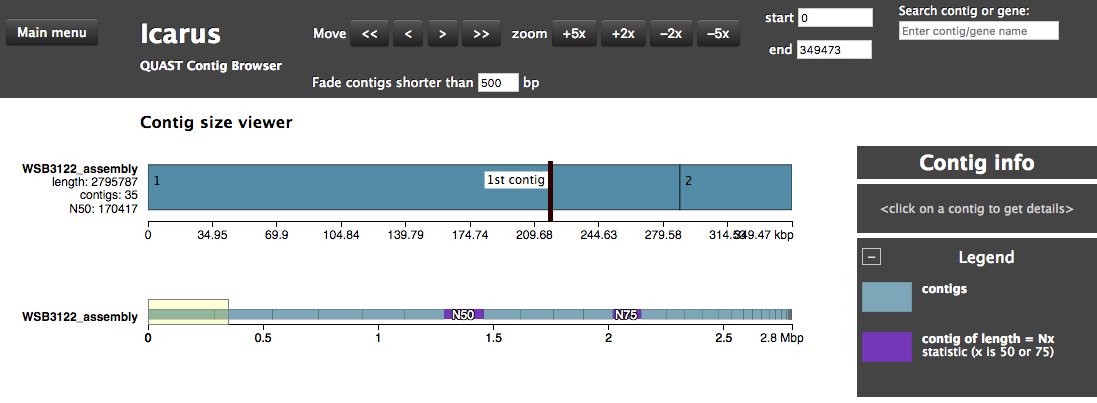
Assembly QC with QUAST
The assembly QC is performed with QUAST. It reports multiple metrics including number of contigs, N50, lengths etc in form of an html report. It further creates an HTML file with integrated contig viewer (Icarus).
Output directory: {sample_id}/QUAST
icarus.html- QUAST’s contig browser as HTML
report.html- QUAST assembly QC as HTML report
Annotation with Prokka
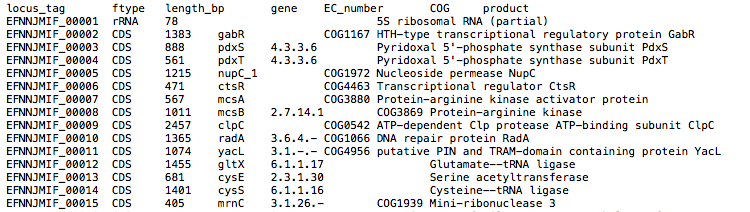
The assembly is annotated with Prokka which acts as frontend for several annotation tools and includes rRNA and ORF predictions. See its documentation for a full description of all output files.
Output directory: {sample_id}/{sample_id}_annotation
Report
Some pipeline results are visualised by MultiQC, which is a visualisation tool that generates a single HTML report summarising all samples in your project. Further statistics are available in within the report data directory.
The pipeline has special steps which allow the software versions used to be reported in the MultiQC output for future traceability.
Output directory: results/multiqc
Project_multiqc_report.html- MultiQC report - a standalone HTML file that can be viewed in your web browser
Project_multiqc_data/- Directory containing parsed statistics from the different tools used in the pipeline
For more information about how to use MultiQC reports, see http://multiqc.info