nf-core/methylseq
Methylation (Bisulfite-Sequencing) analysis pipeline using Bismark/bwa-meth + MethylDackel or bwa-mem + rastair
Introduction
nf-core/methylseq is a bioinformatics analysis pipeline used for Methylation (Bisulfite) sequencing data. It pre-processes raw data from FastQ inputs, aligns the reads and performs extensive quality-control on the results.
The pipeline is built using Nextflow, a workflow tool to run tasks across multiple compute infrastructures in a very portable manner. It uses Docker / Singularity / Podman / Charliecloud / Apptainer containers making installation trivial and results highly reproducible.
On release, automated continuous integration tests run the pipeline on a full-sized dataset on the AWS cloud infrastructure. This ensures that the pipeline runs on AWS, has sensible resource allocation defaults set to run on real-world datasets, and permits the persistent storage of results to benchmark between pipeline releases and other analysis sources.The results obtained from the full-sized test can be viewed on the nf-core website.
Read more about Bisulfite Sequencing & Three-Base Aligners used in this pipeline here
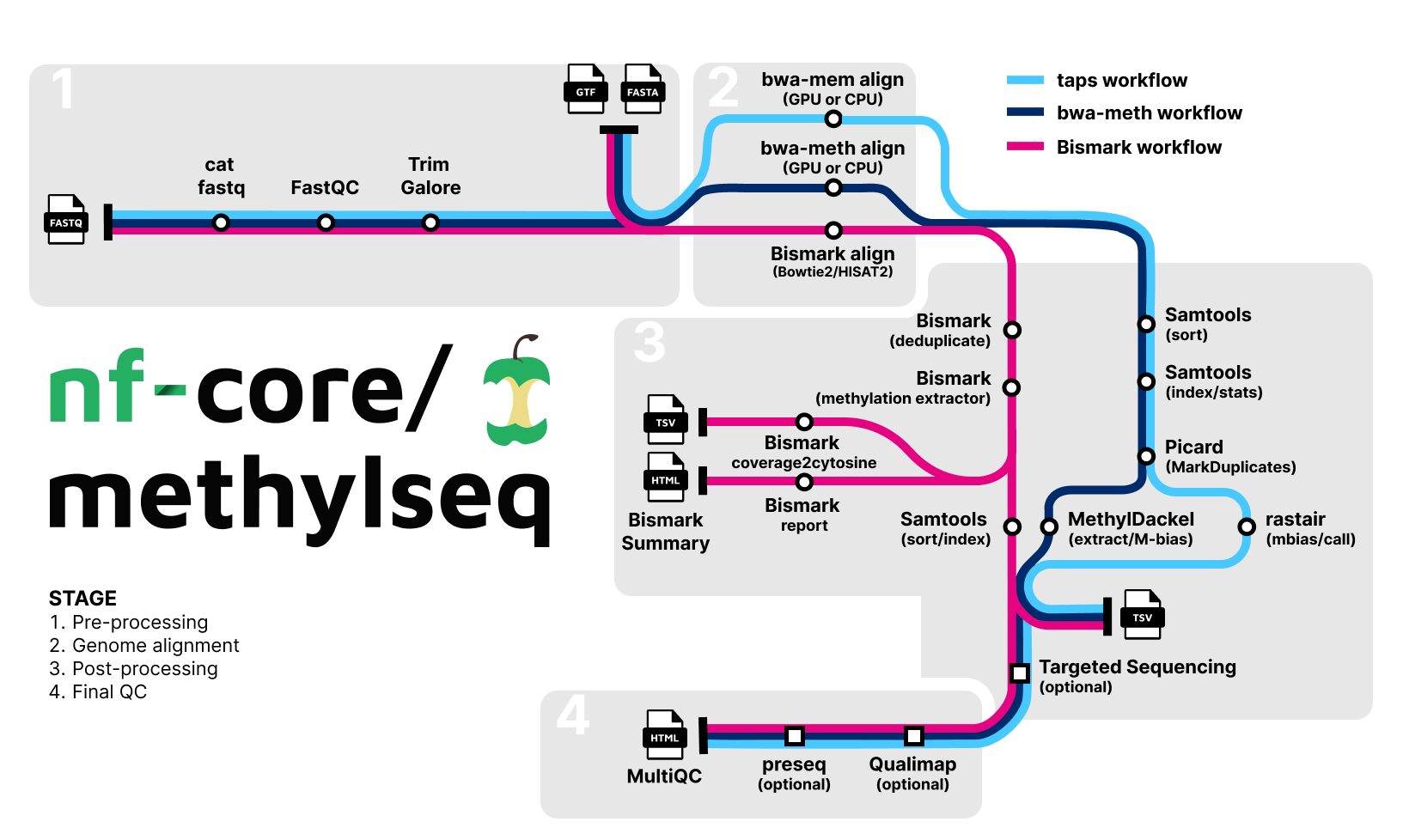
Pipeline Summary
The pipeline allows you to choose between running either Bismark, bwa-meth / MethylDackel or BWA-Mem plus rastair for TAPS data processing. rastair can also be used with bwa-meth aligned reads by setting the aligner to --aligner bwameth and adding the flag --taps.
Choose between workflows by using --aligner bismark (default, uses bowtie2 for alignment), --aligner bismark_hisat, --aligner bwameth or --aligner bwamem. For higher performance, the pipeline can leverage the Parabricks implementation of bwa-meth (fq2bammeth) and the Parabricks implementation of bwa-mem (fq2bammemh), which implement the baseline tools bwa-meth and bwa-mem. To use this option, include the gpu profile along with --aligner bwameth or --aligner bwamem.
Note: For faster CPU runs with BWA-Meth, enable the BWA-MEM2 algorithm using --use_mem2. The GPU pathway (Parabricks) requires -profile gpu and a container runtime (Docker, Singularity, or Podman); Conda/Mamba are not supported for the GPU module.
| Step | Bismark workflow | bwa-meth workflow | bwa-mem + TAPS workflow |
|---|---|---|---|
| Generate Reference Genome Index (optional) | Bismark | bwa-meth | bwa index |
| Merge re-sequenced FastQ files | cat | cat | cat |
| Raw data QC | FastQC | FastQC | FastQC |
| Adapter sequence trimming | Trim Galore! | Trim Galore! | Trim Galore! |
| Align Reads | Bismark (bowtie2/hisat2) | bwa-meth | bwa mem |
| Deduplicate Alignments | Bismark | Picard MarkDuplicates | Picard MarkDuplicates |
| Extract methylation calls | Bismark | MethylDackel | TAPS subworkflow (rastair) |
| Sample report | Bismark | - | - |
| Summary Report | Bismark | - | - |
| Alignment QC | Qualimap (optional) | Qualimap (optional) | Qualimap (optional) |
| Sample complexity | Preseq (optional) | Preseq (optional) | Preseq (optional) |
| Project Report | MultiQC | MultiQC | MultiQC |
Optional targeted sequencing analysis is available via --run_targeted_sequencing and --target_regions_file; see the usage documentation for details.
Usage
If you are new to Nextflow and nf-core, please refer to this page on how to set-up Nextflow. Make sure to test your setup with -profile test before running the workflow on actual data.
First, prepare a samplesheet with your input data that looks as follows:
samplesheet.csv:
sample,fastq_1,fastq_2,genome
SRR389222_sub1,https://github.com/nf-core/test-datasets/raw/methylseq/testdata/SRR389222_sub1.fastq.gz,,
SRR389222_sub2,https://github.com/nf-core/test-datasets/raw/methylseq/testdata/SRR389222_sub2.fastq.gz,,
SRR389222_sub3,https://github.com/nf-core/test-datasets/raw/methylseq/testdata/SRR389222_sub3.fastq.gz,,
Ecoli_10K_methylated,https://github.com/nf-core/test-datasets/raw/methylseq/testdata/Ecoli_10K_methylated_R1.fastq.gz,https://github.com/nf-core/test-datasets/raw/methylseq/testdata/Ecoli_10K_methylated_R2.fastq.gz,Each row represents a fastq file (single-end) or a pair of fastq files (paired end).
Now, you can run the pipeline using default parameters as:
nextflow run nf-core/methylseq --input samplesheet.csv --outdir <OUTDIR> --genome GRCh37 -profile <docker/singularity/podman/shifter/charliecloud/conda/institute>Please provide pipeline parameters via the CLI or Nextflow -params-file option. Custom config files including those provided by the -c Nextflow option can be used to provide any configuration except for parameters; see docs.
For more details and further functionality, please refer to the usage documentation and the parameter documentation.
Pipeline output
To see the results of an example test run with a full size dataset refer to the results tab on the nf-core website pipeline page. For more details about the output files and reports, please refer to the output documentation.
Credits
nf-core/methylseq was originally written by Phil Ewels (@ewels), and Sateesh Peri (@sateeshperi) is its active maintainer.
We thank the following people for their extensive assistance in the development of this pipeline:
- Felix Krueger (@FelixKrueger)
- Edmund Miller (@EMiller88)
- Rickard Hammarén (@Hammarn)
- Alexander Peltzer (@apeltzer)
- Patrick Hüther (@phue)
- Maxime U Garcia (@maxulysse)
Contributions and Support
If you would like to contribute to this pipeline, please see the contributing guidelines.
For further information or help, don’t hesitate to get in touch on the Slack #methylseq channel (you can join with this invite).
Citations
If you use nf-core/methylseq for your analysis, please cite it using the following doi: 10.5281/zenodo.1343417
An extensive list of references for the tools used by the pipeline can be found in the CITATIONS.md file.
You can cite the nf-core publication as follows:
The nf-core framework for community-curated bioinformatics pipelines.
Philip Ewels, Alexander Peltzer, Sven Fillinger, Harshil Patel, Johannes Alneberg, Andreas Wilm, Maxime Ulysse Garcia, Paolo Di Tommaso & Sven Nahnsen.
Nat Biotechnol. 2020 Feb 13. doi: 10.1038/s41587-020-0439-x.