nf-core/mhcquant
Identify and quantify MHC eluted peptides from mass spectrometry raw data
1.6.0). The latest
stable release is
3.0.0
.
Introduction
nfcore/mhcquant is a bioinformatics analysis pipeline used for quantitative processing of data dependent (DDA) peptidomics data.
It was specifically designed to analyse immunopeptidomics data, which deals with the analysis of affinity purified, unspecifically cleaved peptides that have recently been discussed intensively in the context of cancer vaccines.
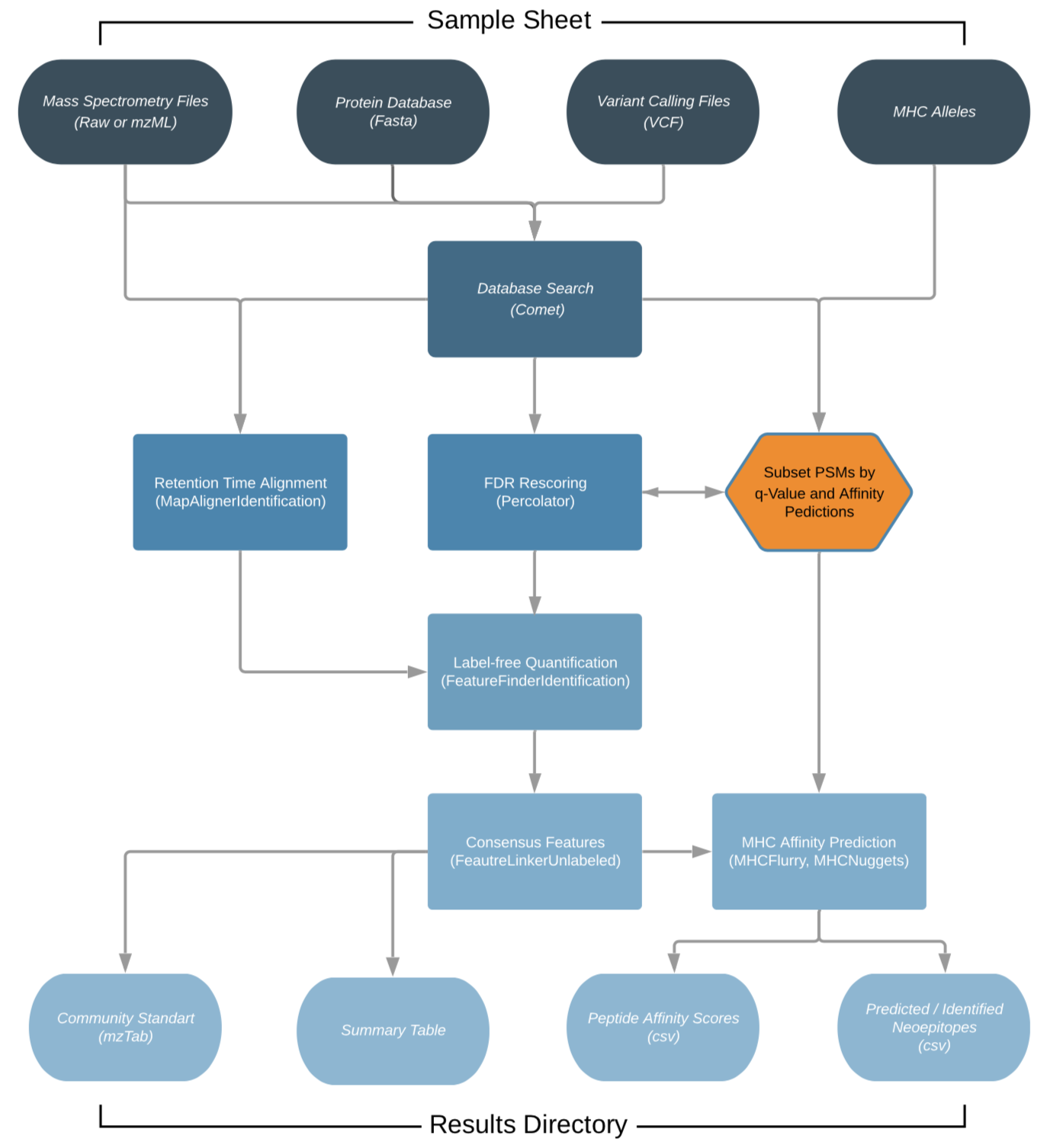
The workflow is based on the OpenMS C++ framework for computational mass spectrometry. RAW files (mzML) serve as inputs and a database search (Comet) is performed based on a given input protein database. FDR rescoring is applied using Percolator based on a competitive target-decoy approach (reversed decoys). For label free quantification all input files undergo identification based retention time alignment (MapAlignerIdentification), and targeted feature extraction matching ids between runs (FeatureFinderIdentification). In addition, a variant calling file (vcf) can be specified to translate variants into proteins that will be included in the database search and binding predictions on specified alleles (alleles.tsv) using MHCFlurry (Class 1) or MHCNugget (Class 2) can be directly run on the output peptide lists. Moreover, if a vcf file was specified, neoepitopes will automatically be determined and binding predictions can also directly be predicted for them.
The pipeline is built using Nextflow, a workflow tool to run tasks across multiple compute infrastructures in a very portable manner. It comes with docker containers making installation trivial and results highly reproducible.
Quick Start
-
Install
nextflow -
Install either
DockerorSingularityfor full pipeline reproducibility (please only useCondaas a last resort; see docs) -
Download the pipeline and test it on a minimal dataset with a single command:
nextflow run nf-core/mhcquant -profile test,<docker/singularity/conda/institute>Please check nf-core/configs to see if a custom config file to run nf-core pipelines already exists for your Institute. If so, you can simply use
-profile <institute>in your command. This will enable eitherdockerorsingularityand set the appropriate execution settings for your local compute environment. -
Start running your own analysis!
nextflow run nf-core/mhcquant --input 'samples.tsv' --fasta 'SWISSPROT_2020.fasta' --allele_sheet 'alleles.tsv' --predict_class_1 --refine_fdr_on_predicted_subset -profile standard,dockerSee usage docs for all of the available options when running the pipeline.
Documentation
The nf-core/mhcquant pipeline comes with documentation about the pipeline which you can read at https://nf-co.re/mhcquant.
Credits
nf-core/mhcquant was originally written by Leon Bichmann, Lukas Heumos and Alexander Peltzer.
Contributions and Support
If you would like to contribute to this pipeline, please see the contributing guidelines.
For further information or help, don’t hesitate to get in touch on the Slack #mhcquant channel (you can join with this invite).
Citation
If you use nf-core/mhcquant for your analysis, please cite:
MHCquant: Automated and Reproducible Data Analysis for Immunopeptidomics
Leon Bichmann, Annika Nelde, Michael Ghosh, Lukas Heumos, Christopher Mohr, Alexander Peltzer, Leon Kuchenbecker, Timo Sachsenberg, Juliane S. Walz, Stefan Stevanović, Hans-Georg Rammensee & Oliver Kohlbacher
Journal of Proteome Research 2019 18 (11), 3876-3884 DOI: 10.1021/acs.jproteome.9b00313
You can cite the nf-core publication as follows:
The nf-core framework for community-curated bioinformatics pipelines.
Philip Ewels, Alexander Peltzer, Sven Fillinger, Harshil Patel, Johannes Alneberg, Andreas Wilm, Maxime Ulysse Garcia, Paolo Di Tommaso & Sven Nahnsen.
Nat Biotechnol. 2020 Feb 13. doi: 10.1038/s41587-020-0439-x. ReadCube: Full Access Link