nf-core/multiplesequencealign
A pipeline to run and systematically evaluate Multiple Sequence Alignment (MSA) methods.
Introduction
Use nf-core/multiplesequencealign to:
- Deploy one (or many) of the most popular Multiple Sequence Alignment (MSA) tools.
- Benchmark MSA tools (and their inputs) using various metrics.
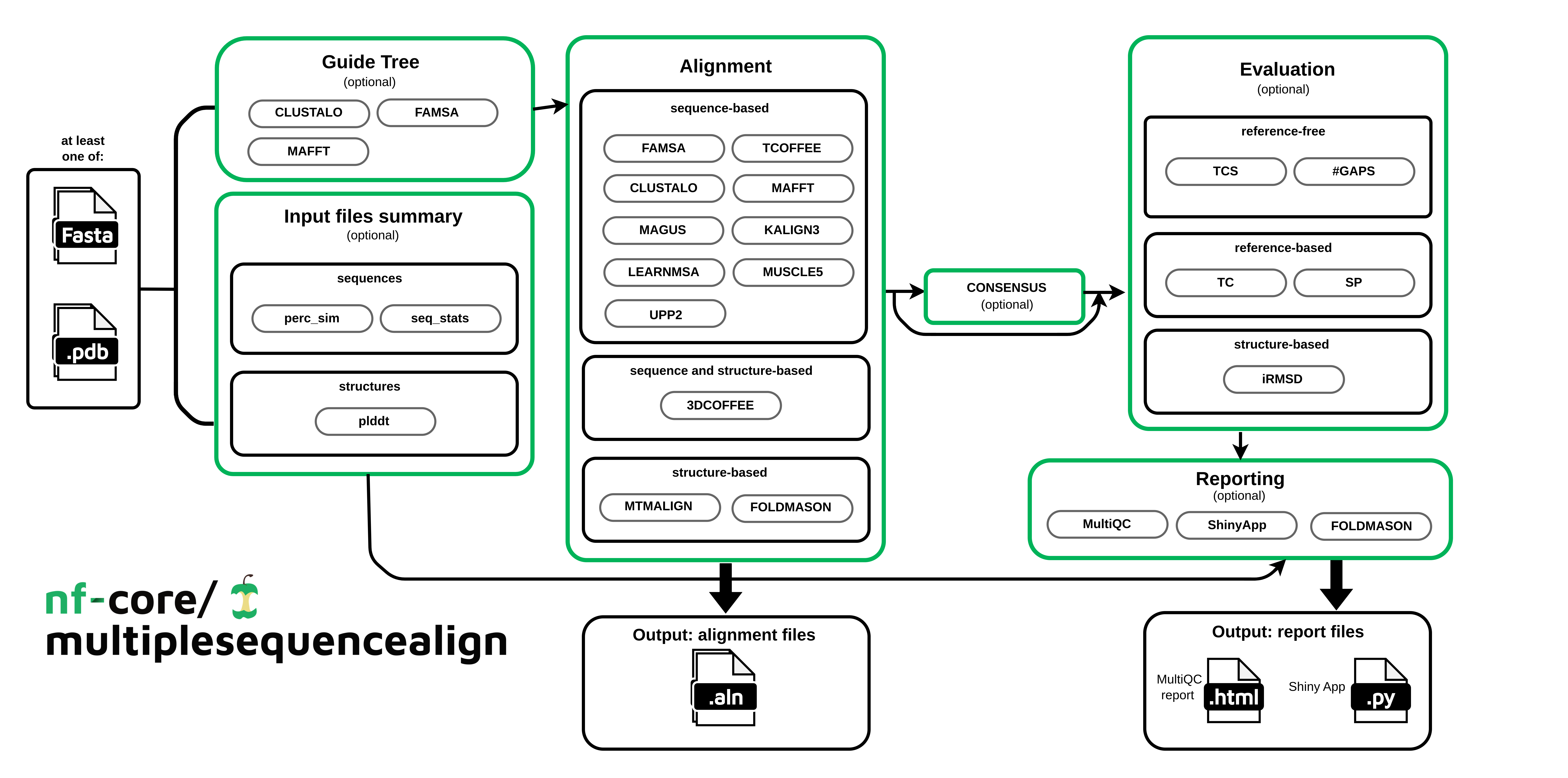
Main steps:
Inputs summary (Optional)
Computation of summary statistics on the input files (e.g., average sequence similarity across the input sequences, their length, pLDDT extraction if available).
Guide Tree (Optional)
Renders a guide tree with a chosen tool (list available in usage). Some aligners use guide trees to define the order in which the sequences are aligned.
Align (Required)
Aligns the sequences with a chosen tool (list available in usage).
Evaluate (Optional)
Evaluates the generated alignments with different metrics: Sum Of Pairs (SoP), Total Column score (TC), iRMSD, Total Consistency Score (TCS), etc.
Report(Optional)
Reports the collected information of the runs in a Shiny app and a summary table in MultiQC. Optionally, it can also render the Foldmason MSA visualization in HTML format.
More introductory material: bytesize talk, nextflow summit talk, poster.
Usage
If you are new to Nextflow and nf-core, please refer to this page on how to set-up Nextflow. Make sure to test your setup with -profile test before running the workflow on actual data.
Quick start - test run
To get a feeling of what the pipeline does, run:
(You don’t need to download or provide any file, try it!)
nextflow run nf-core/multiplesequencealign \
-profile test_tiny,docker \
--outdir resultsand if you want to see how a more complete run looks like, you can try:
nextflow run nf-core/multiplesequencealign \
-profile test,docker \
--outdir resultsHow to set up an easy run:
We have a lot more of use cases examples under FAQs
Input data
You can provide either (or both) a fasta file or a set of protein structures.
Alternatively, you can provide a samplesheet and a toolsheet.
See below how to provide them.
Find some example input data here
CASE 1: One input dataset, one tool.
If you only have one dataset and want to align it using one specific MSA tool (e.g. FAMSA or FOLDMASON), you can run the pipeline with one single command.
Is your input a fasta file (example)? Then:
nextflow run nf-core/multiplesequencealign \
-profile easy_deploy,docker \
--seqs <YOUR_FASTA.fa> \
--aligner FAMSA \
--outdir outdirIs your input a directory where your PDB files are stored (example)? Then:
nextflow run nf-core/multiplesequencealign \
-profile easy_deploy,docker \
--pdbs_dir <PATH_TO_YOUR_PDB_DIR> \
--aligner FOLDMASON \
--outdir outdirFAQ: Which are the available tools I can use?
Check the list here: available tools.FAQ: Can I use both --seqs and --pdbs_dir?
Yes, go for it! This might be useful if you want a structural evaluation of a sequence-based aligner for instance.FAQ: Can I specify also which guidetree to use?
Yes, use the--tree flag. More info: usage and parameters.
FAQ: Can I specify the arguments of the tools (tree and aligner)?
Yes, use the--args_tree and --args_aligner flags. More info: usage and parameters.
CASE 2: Multiple datasets, multiple tools.
nextflow run nf-core/multiplesequencealign \
-profile test,docker \
--input <samplesheet.csv> \
--tools <toolsheet.csv> \
--outdir outdirYou need 2 input files:
- samplesheet (your datasets)
- toolsheet (which tools you want to use).
What is a samplesheet?
The sample sheet defines the input datasets (sequences, structures, etc.) that the pipeline will process.A minimal version:
id,fasta
seatoxin,seatoxin.fa
toxin,toxin.faA more complete one:
id,fasta,reference,optional_data
seatoxin,seatoxin.fa,seatoxin-ref.fa,seatoxin_structures
toxin,toxin.fa,toxin-ref.fa,toxin_structuresEach row represents a set of sequences (in this case the seatoxin and toxin protein families) to be aligned and the associated (if available) reference alignments and dependency files (this can be anything from protein structure or any other information you would want to use in your favourite MSA tool).
Please check: usage.
The only required input is the id column and either fasta or optional_data.
What is a toolsheet?
The toolsheet specifies which combination of tools will be deployed and benchmarked in the pipeline.Each line defines a combination of guide tree and multiple sequence aligner to run with the respective arguments to be used.
The only required field is aligner. The fields tree, args_tree and args_aligner are optional and can be left empty.
A minimal version:
tree,args_tree,aligner,args_aligner
,,FAMSA,This will run the FAMSA aligner.
A more complex one:
tree,args_tree,aligner,args_aligner
FAMSA, -gt upgma -medoidtree, FAMSA,
, ,TCOFFEE,
FAMSA,,REGRESSIVE,This will run, in parallel:
- the FAMSA guidetree with the arguments -gt upgma -medoidtree. This guidetree is then used as input for the FAMSA aligner.
- the TCOFFEE aligner
- the FAMSA guidetree with default arguments. This guidetree is then used as input for the REGRESSIVE aligner.
Please check: usage.
The only required input is aligner.
For more details on more advanced runs: usage documentation and the parameter documentation.
Please provide pipeline parameters via the CLI or Nextflow -params-file option. Custom config files including those provided by the -c Nextflow option can be used to provide any configuration except for parameters; see docs.
Pipeline resources
Which resources is the pipeline using? You can find the default resources used in base.config.
If you are using specific profiles, e.g. test, these will overwrite the defaults.
If you want to modify the needed resources, please refer usage.
Pipeline output
Example results: results tab on the nf-core website pipeline page. For more details: output documentation.
Extending the pipeline
For details on how to add your favourite guide tree, MSA or evaluation step in nf-core/multiplesequencealign please refer to the extending documentation.
Credits
nf-core/multiplesequencealign was originally written by Luisa Santus (@luisas) and Jose Espinosa-Carrasco (@JoseEspinosa) from The Comparative Bioinformatics Group at The Centre for Genomic Regulation, Spain.
The following people have significantly contributed to the development of the pipeline and its modules: Leon Rauschning (@lrauschning), Alessio Vignoli (@alessiovignoli), Igor Trujnara (@itrujnara) and Leila Mansouri (@l-mansouri).
Contributions and Support
If you would like to contribute to this pipeline, please see the contributing guidelines.
For further information or help, don’t hesitate to get in touch on the Slack #multiplesequencealign channel (you can join with this invite).
Citations
If you use nf-core/multiplesequencealign for your analysis, please cite it using the following doi: 10.5281/zenodo.13889386
An extensive list of references for the tools used by the pipeline can be found in the CITATIONS.md file.
You can cite the nf-core publication as follows:
The nf-core framework for community-curated bioinformatics pipelines.
Philip Ewels, Alexander Peltzer, Sven Fillinger, Harshil Patel, Johannes Alneberg, Andreas Wilm, Maxime Ulysse Garcia, Paolo Di Tommaso & Sven Nahnsen.
Nat Biotechnol. 2020 Feb 13. doi: 10.1038/s41587-020-0439-x.