nf-core/quantms
Quantitative mass spectrometry workflow. Currently supports proteomics experiments with complex experimental designs for DDA-LFQ, DDA-Isobaric and DIA-LFQ quantification.
1.0). The latest
stable release is
1.2.0
.
This pipeline is archived and no longer maintained.
Archived pipelines can still be used, but may be outdated and will not receive bugfixes.
Introduction
nf-core/quantms is a bioinformatics best-practice analysis pipeline for Quantitative Mass Spectrometry (MS). Currently, the workflow supports three major MS-based analytical methods: (i) Data dependant acquisition (DDA) label-free and Isobaric quantitation (e.g. TMT, iTRAQ); (ii) Data independent acquisition (DIA) label-free quantification (for details see our in-depth documentation on quantms).
The pipeline is built using Nextflow, a workflow tool to run tasks across multiple compute infrastructures in a very portable manner. It uses Docker/Singularity containers making installation trivial and results highly reproducible. The Nextflow DSL2 implementation of this pipeline uses one container per process which makes it much easier to maintain and update software dependencies. Where possible, these processes have been submitted to and installed from nf-core/modules in order to make them available to all nf-core pipelines, and to everyone within the Nextflow community!
On release, automated continuous integration tests run the pipeline on a full-sized dataset on the AWS cloud infrastructure. This ensures that the pipeline runs on AWS, has sensible resource allocation defaults set to run on real-world datasets, and permits the persistent storage of results to benchmark between pipeline releases and other analysis sources. The results obtained from the full-sized test can be viewed on the nf-core website. This gives you a hint on which reports and file types are produced by the pipeline in a standard run. The automatic continuous integration tests evaluate different workflows, including the peptide identification, quantification for LFQ, LFQ-DIA, and TMT test datasets.
Pipeline summary
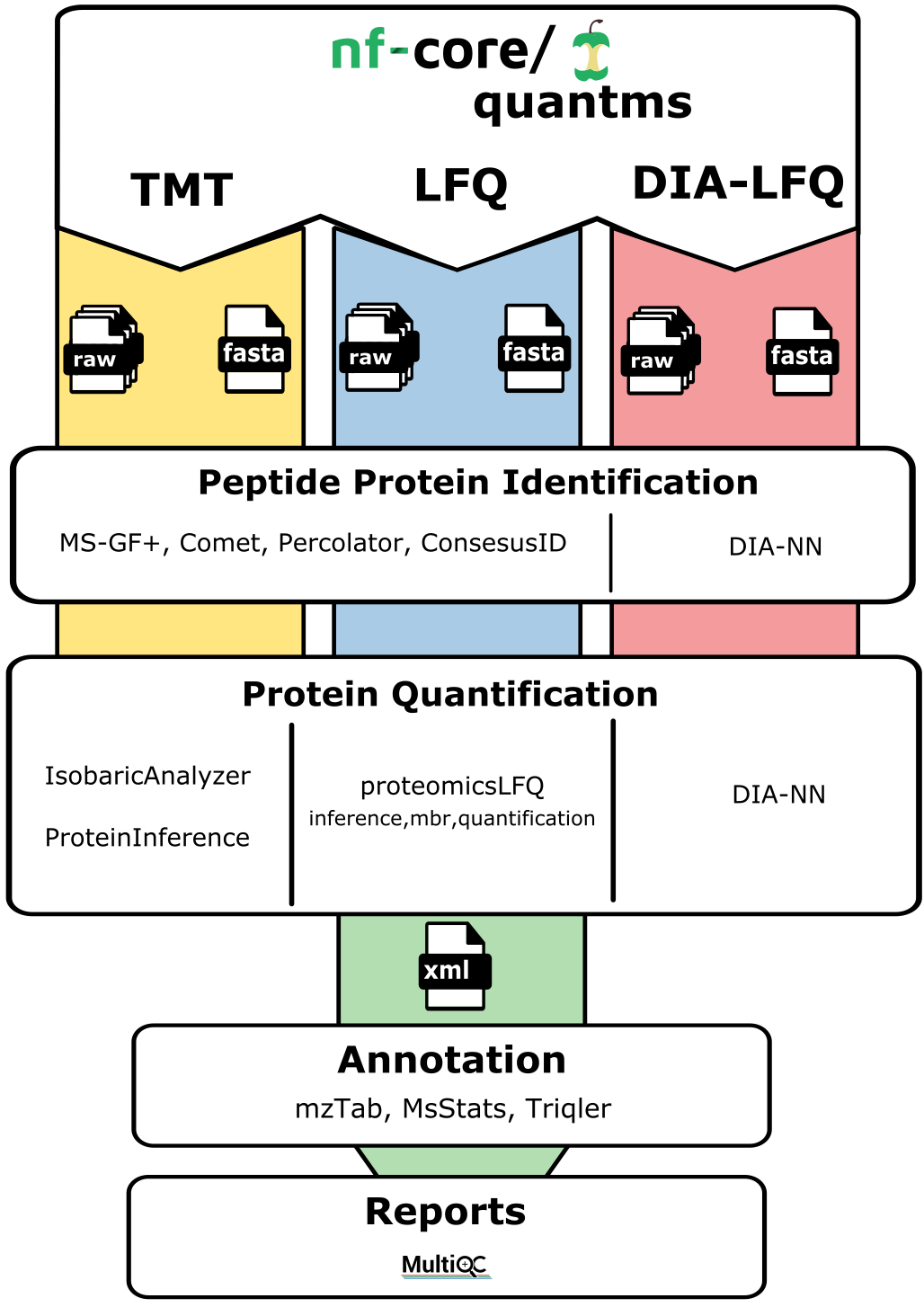
The quantms allows uses to perform analysis in three main type of analytical MS-based quantitative methods: DDA-LFQ, DDA-ISO, DIA-LFQ. Each of these workflows share some processes but also includes their own steps. In summary:
DDA-LFQ:
- RAW file conversion to mzML (
thermorawfileparser) - Peptide identification using
cometand/ormsgf+ - Re-scoring peptide identifications
percolator - Peptide identification FDR
openms fdr tool - Modification localization
luciphor - Quantification: Feature detection
proteomicsLFQ - Protein inference and quantification
proteomicsLFQ - QC report generation
pmultiqc - Normalization, imputation, significance testing with
MSstats
DDA-ISO:
- RAW file conversion to mzML (
thermorawfileparser) - Peptide identification using
cometand/ormsgf+ - Re-scoring peptide identifications
percolator - Peptide identification FDR
openms fdr tool - Modification localization
luciphor - Extracts and normalizes isobaric labeling
IsobaricAnalyzer - Protein inference
ProteinInferenceorEpifanyfor bayesian inference. - Protein Quantification
ProteinQuantifier - QC report generation
pmultiqc - Normalization, imputation, significance testing with
MSstats
DIA-LFQ:
- RAW file conversion to mzML (
thermorawfileparser) - DIA-NN analysis
dia-nn - Generation of output files (msstats)
- QC reports generation
pmultiqc
Functionality overview
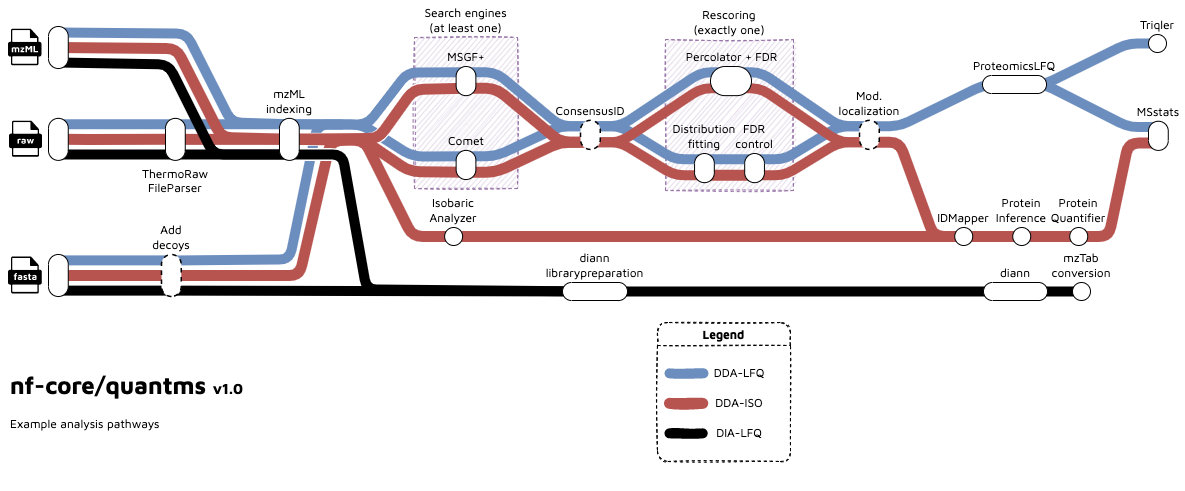
A graphical overview of suggested routes through the pipeline depending on context can be seen below.
Quick Start
-
Install
Nextflow(>=21.10.3) -
Install any of
Docker,Singularity,Podman,ShifterorCharliecloudfor full pipeline reproducibility (please only useCondaas a last resort; see docs) -
Download the pipeline and test it on a minimal dataset with a single command:
nextflow run nf-core/quantms -profile test,YOURPROFILE --input project.sdrf.tsv --database protein.fasta --outdir <OUTDIR>Note that some form of configuration will be needed so that Nextflow knows how to fetch the required software. This is usually done in the form of a config profile (
YOURPROFILEin the example command above). You can chain multiple config profiles in a comma-separated string.-
The pipeline comes with config profiles called
docker,singularity,podman,shifter,charliecloudandcondawhich instruct the pipeline to use the named tool for software management. For example,-profile test,docker. -
Please check nf-core/configs to see if a custom config file to run nf-core pipelines already exists for your Institute. If so, you can simply use
-profile <institute>in your command. This will enable eitherdockerorsingularityand set the appropriate execution settings for your local compute environment. -
If you are using
singularityand are persistently observing issues downloading Singularity images directly due to timeout or network issues, then you can use the--singularity_pull_docker_containerparameter to pull and convert the Docker image instead. Alternatively, you can use thenf-core downloadcommand to download images first, before running the pipeline. Setting theNXF_SINGULARITY_CACHEDIRorsingularity.cacheDirNextflow options enables you to store and re-use the images from a central location for future pipeline runs. -
If you are using
conda, it is highly recommended to use theNXF_CONDA_CACHEDIRorconda.cacheDirsettings to store the environments in a central location for future pipeline runs.
- The pipeline comes with config profiles called
docker,singularity,podman,shifter,charliecloudandcondawhich instruct the pipeline to use the named tool for software management. For example,-profile test,docker. - Please check nf-core/configs to see if a custom config file to run nf-core pipelines already exists for your Institute. If so, you can simply use
-profile <institute>in your command. This will enable eitherdockerorsingularityand set the appropriate execution settings for your local compute environment. - If you are using
singularity, please use thenf-core downloadcommand to download images first, before running the pipeline. Setting theNXF_SINGULARITY_CACHEDIRorsingularity.cacheDirNextflow options enables you to store and re-use the images from a central location for future pipeline runs. - If you are using
conda, it is highly recommended to use theNXF_CONDA_CACHEDIRorconda.cacheDirsettings to store the environments in a central location for future pipeline runs.
-
-
Start running your own analysis!
nextflow run nf-core/quantms -profile <docker/singularity/podman/shifter/charliecloud/conda/institute> --input project.sdrf.tsv --database database.fasta --outdir <OUTDIR>
Documentation
The nf-core/quantms pipeline comes with a stand-alone full documentation including examples, benchmarks, and detailed explanation about the data analysis of proteomics data using quantms. In addition, quickstart documentation of the pipeline can be found in: usage, parameters and output.
Credits
nf-core/quantms was originally written by: Chengxin Dai (@daichengxin), Julianus Pfeuffer (@jpfeuffer) and Yasset Perez-Riverol (@ypriverol).
We thank the following people for their extensive assistance in the development of this pipeline:
- Timo Sachsenberg (@timosachsenberg)
- Wang Hong (@WangHong007)
Contributions and Support
If you would like to contribute to this pipeline, please see the contributing guidelines.
For further information or help, don’t hesitate to get in touch on the Slack #quantms channel (you can join with this invite). In addition, users can get in touch using our discussion forum
Citations
An extensive list of references for the tools used by the pipeline can be found in the CITATIONS.md file.
You can cite the nf-core publication as follows:
The nf-core framework for community-curated bioinformatics pipelines.
Philip Ewels, Alexander Peltzer, Sven Fillinger, Harshil Patel, Johannes Alneberg, Andreas Wilm, Maxime Ulysse Garcia, Paolo Di Tommaso & Sven Nahnsen.
Nat Biotechnol. 2020 Feb 13. doi: 10.1038/s41587-020-0439-x.