nf-core/airrflow
B-cell and T-cell Adaptive Immune Receptor Repertoire (AIRR) sequencing analysis pipeline using the Immcantation framework
3.3.0). The latest
stable release is
4.3.1
.
Introduction
This document describes the output produced by the pipeline. Most of the plots are taken from the MultiQC report, which summarises results at the end of the pipeline.
The directories listed below will be created in the results directory after the pipeline has finished. All paths are relative to the top-level results directory.
Pipeline overview
The pipeline is built using Nextflow and processes data using the following steps:
- QC and sequence assembly (bulk only)
- FastP - read quality control, adapter trimming and read clipping.
- Filter by sequence quality - filter sequences by base quality.
- Mask primers - Mask amplicon primers.
- Pair mates - Pair read mates.
- Cluster sets - Cluster sequences according to similarity.
- Build consensus - Build consensus of sequences with the same UMI barcode.
- Re-pair mates - Re-pairing sequence mates.
- Assemble mates - Assemble sequence mates.
- Remove duplicates - Remove and annotate read duplicates.
- Filter sequences for at least 2 representative Filter sequences that do not have at least 2 duplicates.
- FastQC - read quality control post-assembly
- VDJ annotation - Assign genes and clonotyping
- Bulk QC filtering
- Single cell QC
- Clonal analysis
- Find clonal threshold
- SCOPer define clones - Defining clonal B-cell or T-cell groups
- Dowser lineage reconstruction - Clonal lineage reconstruction.
- Repertoire analysis - Repertoire analysis and comparison.
- Report file size - Log parsing.
- Log parsing - Log parsing.
- Databases - Downloaded databases.
- MultiQC - MultiQC report.
- Pipeline information - Pipeline information
Sequence assembly
NB: If using the sans-UMI subworkflow by specifying
umi_length=0, the presto directory ordering numbers will differ e.g., mate pair assembly results will be output topresto/01-assemblepairs/<sampleID>as this will be the first presto step.
Fastp
Output files
fastp/<sample_id>/*.fastp.html: Fast report containing quality metrics for the mated and quality filtered reads.*.fastp.json: Zip archive containing the FastQC report, tab-delimited data file and plot images for the mated and quality filtered reads.*.fastp.log: Fastp
fastp gives general quality metrics about your sequenced reads, as well as allows filtering reads by quality, trimming adapters and clipping reads at 5’ or 3’ ends. It provides information about the quality score distribution across your reads, per base sequence content (%A/T/G/C), adapter contamination and overrepresented sequences. For further reading and documentation see the fastp documentation.
Filter by sequence quality
Output files
presto/01-filterseq/<sampleID>logs: Raw command logs of the process that will be parsed to generate a report.tabs: Table containing read ID and quality for each of the read files.
Filters reads that are below a quality threshold by using the tool FilterSeq from the pRESTO Immcantation toolset. The default quality threshold is 20.
Mask primers
Output files
presto/02-maskprimers/<sampleID>logs: Raw command logs of the process that will be parsed to generate a report.tabs: Table containing a read ID, the identified matched primer and the error for primer alignment.
Masks primers that are provided in the C-primers and V-primers input files. It uses the tool MaskPrimers of the pRESTO Immcantation toolset.
Pair mates
Output files
presto/03-pairseq/<sampleID>logs: Raw command logs of the process that will be parsed to generate a report.
Pair read mates using PairSeq from the pRESTO Immcantation toolset.
Cluster sets
Output files
presto/04-cluster_sets/<sampleID>logs: Raw command logs of the process that will be parsed to generate a report.tabs: Table containing a read ID, the identified barcode, the cluster id and the number of sequences in the cluster.
Cluster sequences according to similarity, using ClusterSets set. This step is introduced to deal with too low UMI diversity.
Parse clusters
Output files
presto/05-parse_clusters/<sampleID>logs: Raw command logs of the process that will be parsed to generate a report.
Annotate cluster ID as part of the barcode, using Parseheaders copy. This step is introduced to deal with too low UMI diversity.
Build UMI consensus
Output files
presto/06-build_consensus/<sampleID>logs: Raw command logs of the process that will be parsed to generate a report.tabs: Table containing the sequence barcode, number of sequences used to build the consensus (SEQCOUNT), the identified primer (PRIMER), the number of sequences for each primer (PRCOUNT), the primer consensus (PRCONS), the primer frequency (PRFREQ) and the number of sequences used to build the consensus (CONSCOUNT).
Build sequence consensus from all sequences that were annotated to have the same UMI. Uses BuildConsensus from the pRESTO Immcantation toolset.
Re-pair mates
Output files
presto/07-pairseq_postconsensus/<sampleID>logs: Raw command logs of the process that will be parsed to generate a report.
Re-pair read mates using PairSeq from the pRESTO Immcantation toolset.
Assemble mates
Output files
presto/08-assemblepairs/<sampleID>logs: Raw command logs of the process that will be parsed to generate a report.tabs: Parsed log contaning the sequence barcodes and sequence length, bases of the overlap, error of the overlap and p-value.
Assemble read mates using AssemblePairs from the pRESTO Immcantation toolset.
Remove duplicates
Output files
presto/09-collapseseq/<sampleID>logs: Raw command logs of the process that will be parsed to generate a report.tabs: Parsed log containing the sequence barcodes, header information and deduplicate count.
Remove duplicates using CollapseSeq from the pRESTO Immcantation toolset.
Filter sequences for at least 2 representatives
Output files
presto/10-splitseq/<sampleID>logs: Raw command logs of the process that will be parsed to generate a report.
Remove sequences which do not have 2 representative using SplitSeq from the pRESTO Immcantation toolset.
FastQC
Output files
fastqc/postassembly/*_ASSEMBLED_fastqc.html: FastQC report containing quality metrics for the mated and quality filtered reads.*_ASSEMBLED_fastqc.zip: Zip archive containing the FastQC report, tab-delimited data file and plot images for the mated and quality filtered reads.
FastQC gives general quality metrics about your sequenced reads. It provides information about the quality score distribution across your reads, per base sequence content (%A/T/G/C), adapter contamination and overrepresented sequences. For further reading and documentation see the FastQC help pages.
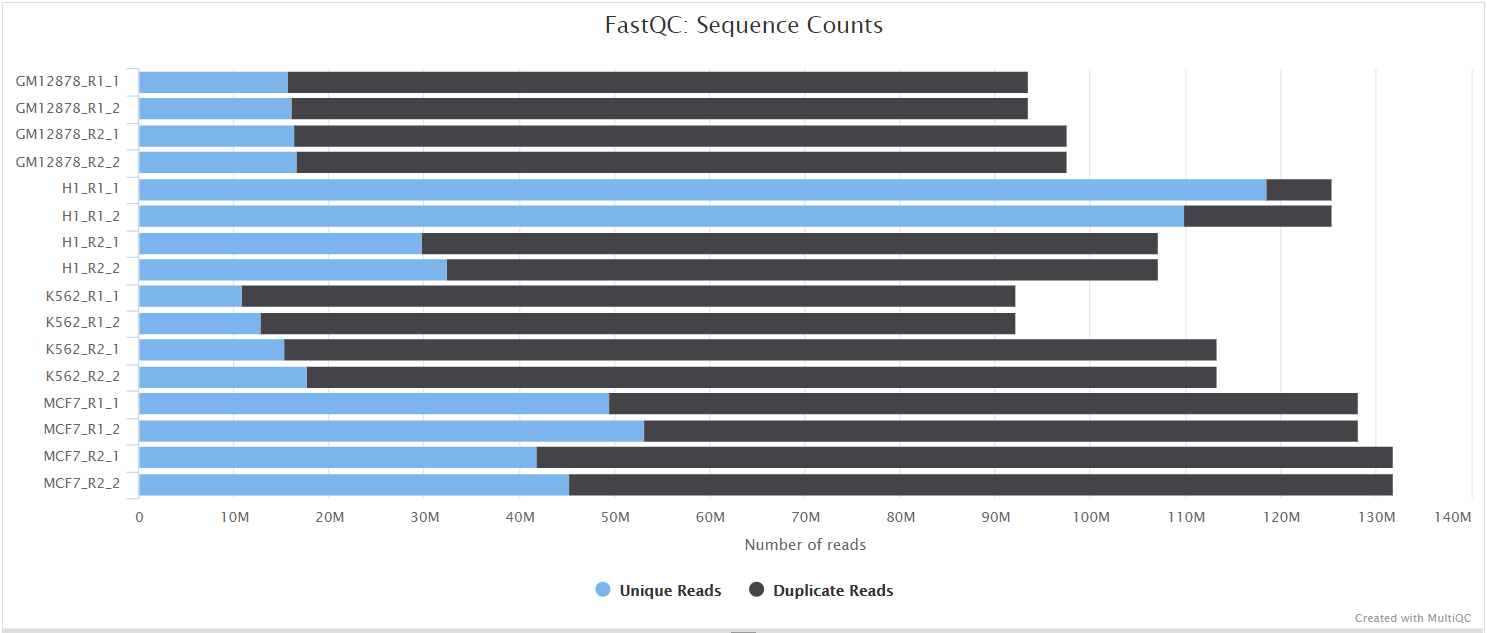
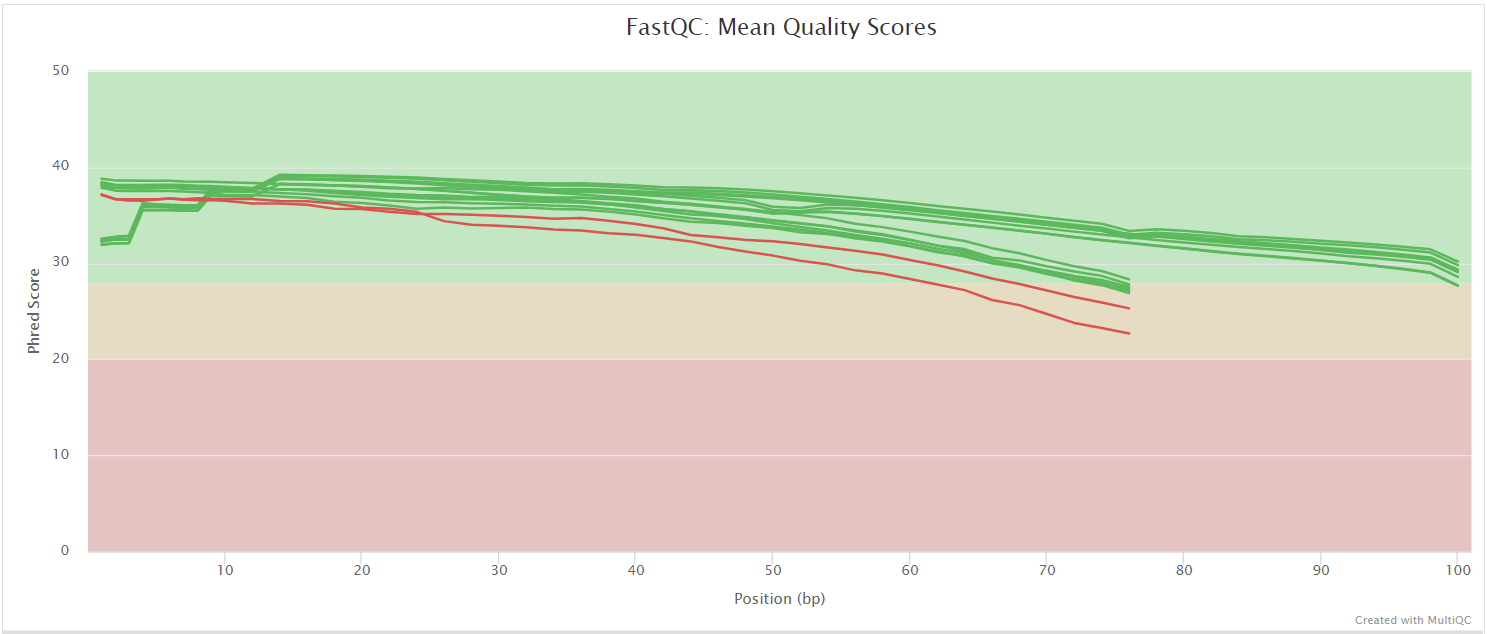
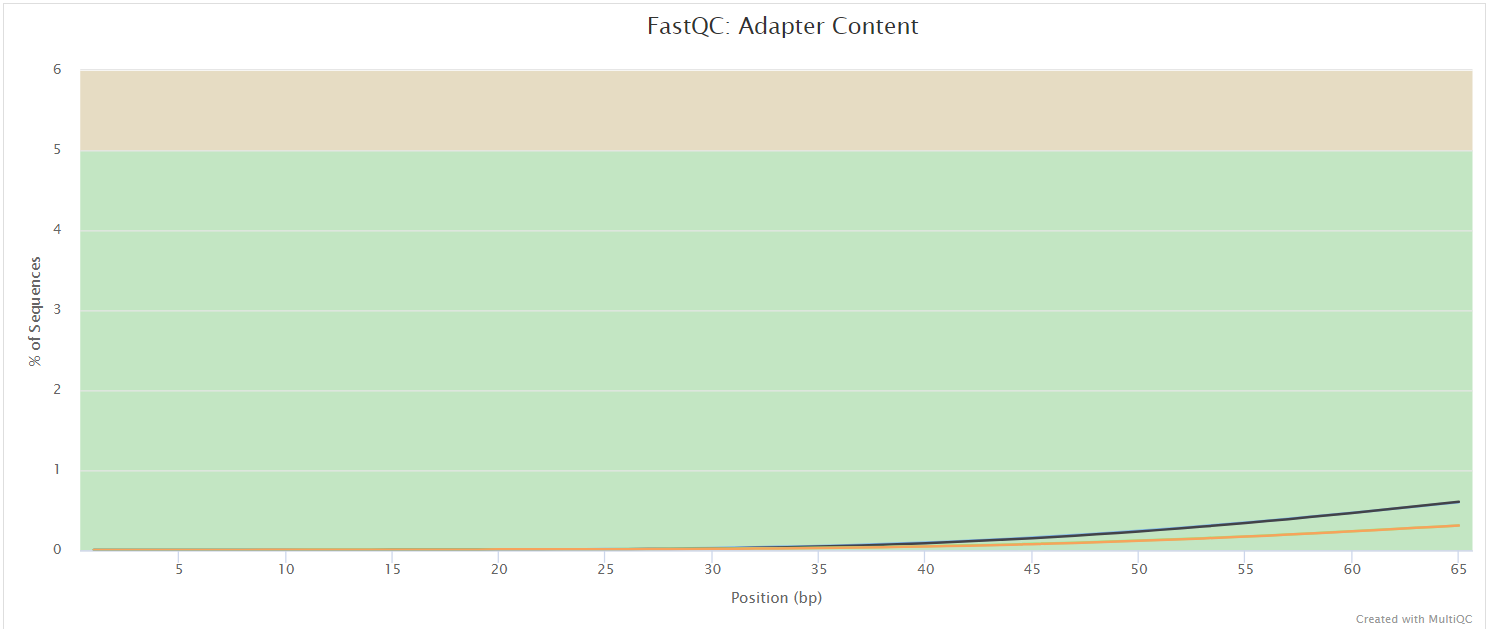
Two sets of FastQC plots are displayed in the MultiQC report: first for the raw untrimmed and unmated reads and secondly for the assembled and QC filtered reads (but before collapsing duplicates). They may contain adapter sequence and potentially regions with low quality.
VDJ annotation
Convert input to fasta (optional)
Output files. Optional.
vdj_annotation/convert-db/<sampleID>*.fasta: The sequences in fasta format.*log.txt: Log of the process that will be parsed to generate a report.
This folder is generated when the input data are AIRR-C formatted rearrangement tables that need to
be reprocessed (--reassign true). For example, 10x Genomics’ airr_rearrangement.tsv files. ConvertDb fasta is used to
generate a .fasta file from the rearrangement table.
Assign genes with Igblast
Output files
vdj_annotation/01-assigngenes/<sampleID>*.fmt7: Igblast results.*.fasta: Igblast results converted to fasta.*log.txt: Log of the process that will be parsed to generate a report.
Assign genes with Igblast, using the IMGT database is performed by the AssignGenes command of the Change-O tool from the Immcantation Framework.
Make database from assigned genes
Output files
vdj_annotation/02-makedb/<sampleID>*log.txt: Log of the process that will be parsed to generate a report.*db-pass.tsv: Rearrangement table in AIRR-C format containing the assigned gene information.
IgBLAST’s results are parsed and standardized with MakeDB to follow the AIRR Community standards for rearrangement data.
Quality filter alignments
Output files
vdj_annotation/03-quality-filter/<sampleID>*log.txt: Log of the process that will be parsed to generate a report.*quality-pass.tsv*: Rearrangement table in AIRR-C format containing the sequences that passed the quality filtering steps.
A table is generated that retains sequences with concordant locus in the v_call and locus fields, with a sequence_alignment with a maximum of 10% of Ns and a length of at least 200 informative nucleotides (not -, . or N).
Removal of non-productive sequences
Output files
vdj_annotation/04-select-productive/<sampleID>*log.txt: Log of the process that will be parsed to generate a report.*productive-T.tsv*: Rearrangement table in AIRR-C format, with only productive sequences.
Non-functional sequences identified with IgBLAST are removed with ParseDb.
Removal of sequences with junction length not multiple of 3
Output files
vdj_annotation/05-select-junction-mod3/<sampleID>*log.txt: Log of the process that will be parsed to generate a report.*junction-pass.tsv*: Rearrangement table in AIRR-C format, with only sequences that have a nucleotide junction length multiple of 3.
Annotate metadata
Output files
vdj_annotation/06-annotate-metadata/<sampleID>*log.txt: Log of the process that will be parsed to generate a report.*meta-pass.tsv*: Rearrangement table in AIRR-C format annotated with metadata provided in the starting metadata sheet.
Bulk QC filtering
Reconstruct germlines
Output files
qc-filtering/bulk-qc-filtering/01-create-germlines/<sampleID>*log.txt: Log of the process that will be parsed to generate a report.*germ-pass.tsv: Rearrangement table in AIRR-C format with an additional field with the reconstructed germline sequence for each sequence.
Reconstructing the germline sequences with the CreateGermlines Immcantation tool.
Chimeric read filtering (optional)
Output files
qc-filtering/bulk-qc-filtering/02-chimera-filter/<sampleID>*log.txt: Log of the process that will be parsed to generate a report.*chimera-pass.tsv: Rearrangement table in AIRR-C format sequences that passed the chimera removal filter.<sampleID>_chimera_report: Report with plots showing the mutation patterns
Mutations patterns in different window sizes are analyzed with functions from the Immcantation R package SHazaM.
Detect contamination (optional)
Output files. Optional.
qc-filtering/bulk-qc-filtering/03-detect_contamination*log.txt: Log of the process that will be parsed to generate a report.*cont-flag.tsv: Rearrangement table in AIRR-C format with sequences that passed the chimera removal filter.all_reps_cont_report: Report.
This folder is genereated when detect_contamination is set to true.
Collapse duplicates
Output files.
qc-filtering/bulk-qc-filtering/04-collapse-duplicates/<sampleID>*log.txt: Log of the process that will be parsed to generate a report.*collapse_report/: Report.repertoires/*collapse-pass.tsv: Rearrangement table in AIRR-C format with duplicated sequences removed.
Single cell QC
Output files.
qc-filtering/single-cell-qc/all_reps_scqc_report*log.txt: Log of the process that will be parsed to generate a report.*all_reps_scqc_report/: Report.*scqc-pass.tsv: Rearrangement table in AIRR-C format with sequences that passed the quality filtering.
Clonal analysis
Find clonal threshold
Output files
clonal_analysis/find_threshold/*log: Log of the process that will be parsed to generate a report.all_reps_dist_report: Reporttables/all_reps_threshold-mean.tsv: Mean of all hamming distance thresholds of the Junction regions as determined by Shazam.tables/all_reps_threshold-summary.tsv: Thresholds for each group of--clonebysamples.
Determining the hamming distance threshold of the junction regions for clonal determination using Shazam when clonal_threshold is set to auto.
SCOPer define clones
Output files
clonal_analysis/define_clones/<subjectID>*log: Log of the process that will be parsed to generate a report.repertoires/<sampleID>_clone-pass.tsv: Rearrangement tables in AIRR-C format with sequences that passed the clonal assignment step. The fieldclone_idcontains the clonal clusters identifiers.tables/: Table in AIRR format containing the assigned gene information and an additional field with the clone id.clonal_abundance.tsvclonal_diversity.tsvclone_sizes_table.tsvnum_clones_table_nosingle.tsvnum_clones_table.tsv
ggplots/: Diversity and abundance plots asggplotobjects.figures/: Clone size, diversity and abundancepngplots.
A similar output folder clonal_analysis/define_clones/all_reps_clone_report is generated for all data, with additional
ggplot objects and png figures showing the convergence between samples.
Assigning clones to the sequences obtained from IgBlast with the scoper::hierarchicalClones Immcantation tool.
Dowser Lineage reconstruction
Output files
clonal_analysis/dowser_lineages/<sampleID>*log: Log of the process that will be parsed to generate a report.<sample1ID>_dowser_report: Report
Reconstructing clonal lineage with IgPhyML and dowser from the Immcantation toolset.
Repertoire analysis
Output files
repertoire_analysis/repertoire_comparison/all_data.tsv: AIRR format table containing the processed sequence information for all subjects.Abundance: contains clonal abundance calculation plots and tables.Diversity: contains diversity calculation plots and tables.V_family: contains V gene and family distribution calculation plots and tables.
Airrflow_report.html: Contains the repertoire comparison results in an html report form: Abundance, Diversity, V gene usage tables and plots. Comparison between treatments and subjects.
Calculation of several repertoire characteristics (diversity, abundance, V gene usage) for comparison between subjects, time points and cell populations. An Rmarkdown report is generated with the Alakazam R package.
Report file size
Output files
report_file_size/file_size_report: Report summarizing the number of sequences after the most important pipeline steps.tables/*tsv: Tables with the number of sequences at each processing step.
Parsing the logs from the previous processes. Summary of the number of sequences left after each of the most important pipeline steps.
Log parsing
Output files
parsed_logs/sequences_table: table summarizing of the number of sequences after the most important pipeline steps.
Parsing the logs from the previous processes. Summary of the number of sequences left after each of the most important pipeline steps.
Databases
Copy of the downloaded IMGT database by the process fetch_databases, used for the gene assignment step.
If databases are provided with --imgtdb_base and --igblast_base this folder will not be present.
MultiQC
Output files
multiqc/multiqc_report.html: a standalone HTML file that can be viewed in your web browser.multiqc_data/: directory containing parsed statistics from the different tools used in the pipeline.multiqc_plots/: directory containing static images from the report in various formats.
MultiQC is a visualization tool that generates a single HTML report summarising all samples in your project. Most of the pipeline QC results are visualised in the report and further statistics are available in the report data directory.
Results generated by MultiQC collate pipeline QC from supported tools e.g. FastQC. The pipeline has special steps which also allow the software versions to be reported in the MultiQC output for future traceability. For more information about how to use MultiQC reports, see http://multiqc.info.
Pipeline information
Output files
pipeline_info/- Reports generated by Nextflow:
execution_report.html,execution_timeline.html,execution_trace.txtandpipeline_dag.dot/pipeline_dag.svg. - Reports generated by the pipeline:
pipeline_report.html,pipeline_report.txtandsoftware_versions.yml. Thepipeline_report*files will only be present if the--email/--email_on_failparameter’s are used when running the pipeline. - Reformatted samplesheet files used as input to the pipeline:
samplesheet.valid.csv. - Parameters used by the pipeline run:
params.json.
- Reports generated by Nextflow:
Nextflow provides excellent functionality for generating various reports relevant to the running and execution of the pipeline. This will allow you to troubleshoot errors with the running of the pipeline, and also provide you with other information such as launch commands, run times and resource usage.