nf-core/pairgenomealign
Pairwise genome comparison pipeline using the LAST software to align a list of query genomes to a target genome, and plot the results
Introduction
nf-core/pairgenomealign is a bioinformatics pipeline that aligns one or more query genomes to a target genome, and plots pairwise representations.
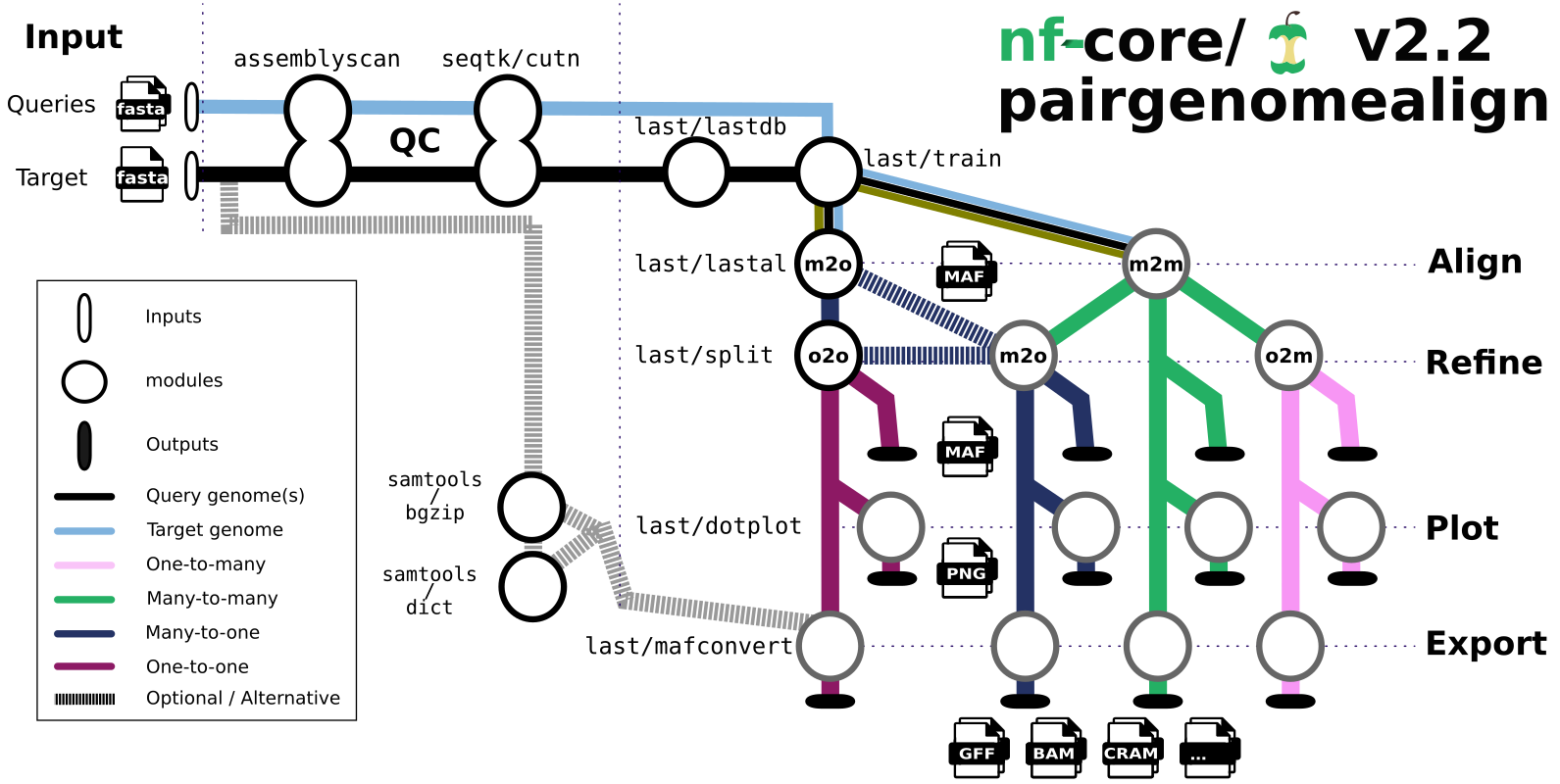
The main steps of the pipeline are:
- Genome QC (
assembly-scan). - Genome indexing (
lastdb). - Genome pairwise alignments (
lastal). - Alignment plotting (
last-dotplot). - Alignment export to various formats with
maf-convert, plusSamtoolsfor SAM/BAM/CRAM.
The pipeline can generate four kinds of outputs, called many-to-many, many-to-one, one-to-many and one-to-one, depending on whether sequences of one genome are allowed match the other genome multiple times or not.
These alignments are output in MAF format, and optional line plot representations are output in PNG format.
Usage
If you are new to Nextflow and nf-core, please refer to this page on how to set-up Nextflow. Make sure to test your setup with -profile test before running the workflow on actual data.
First, prepare a samplesheet with your input data that looks as follows:
samplesheet.csv:
sample,fasta
query_1,path-to-query-genome-file-one.fasta
query_2,path-to-query-genome-file-two.fastaEach row represents a fasta file, this can also contain multiple rows to accomodate multiple query genomes in fasta format.
Now, you can run the pipeline using:
nextflow run nf-core/pairgenomealign \
-profile <docker/singularity/.../institute> \
--target sequencefile.fa \
--input samplesheet.csv \
--outdir <OUTDIR>Please provide pipeline parameters via the CLI or Nextflow -params-file option. Custom config files including those provided by the -c Nextflow option can be used to provide any configuration except for parameters; see docs.
For more details and further functionality, please refer to the usage documentation and the parameter documentation.
Pipeline output
To see the results of an example test run with a full size dataset refer to the results tab on the nf-core website pipeline page. For more details about the output files and reports, please refer to the output documentation.
Credits
nf-core/pairgenomealign was originally written by charles-plessy; the original versions are available at https://github.com/oist/plessy_pairwiseGenomeComparison.
We thank the following people for their extensive assistance in the development of this pipeline:
- Mahdi Mohammed ported the original pipeline to nf-core template 2.14.x.
- Martin Frith, the author of LAST, gave us extensive feedback and advices.
- Michael Mansfield tested the pipeline and provided critical comments.
- Aleksandra Bliznina contributed to the creation of the initial
last/*modules. - Jiashun Miao and Huyen Pham tested the pipeline on vertebrate genomes.
Contributions and Support
If you would like to contribute to this pipeline, please see the contributing guidelines.
For further information or help, don’t hesitate to get in touch on the Slack #pairgenomealign channel (you can join with this invite).
Citations
If you use this pipeline, please cite:
Extreme genome scrambling in marine planktonic Oikopleura dioica cryptic species. Charles Plessy, Michael J. Mansfield, Aleksandra Bliznina, Aki Masunaga, Charlotte West, Yongkai Tan, Andrew W. Liu, Jan Grašič, María Sara del Río Pisula, Gaspar Sánchez-Serna, Marc Fabrega-Torrus, Alfonso Ferrández-Roldán, Vittoria Roncalli, Pavla Navratilova, Eric M. Thompson, Takeshi Onuma, Hiroki Nishida, Cristian Cañestro, Nicholas M. Luscombe. Genome Res. 2024. 34: 426-440; doi: 10.1101/2023.05.09.539028. PubMed ID: 38621828
And also please cite the LAST papers.
An extensive list of references for the tools used by the pipeline can be found in the CITATIONS.md file.
You can cite the nf-core publication as follows:
The nf-core framework for community-curated bioinformatics pipelines.
Philip Ewels, Alexander Peltzer, Sven Fillinger, Harshil Patel, Johannes Alneberg, Andreas Wilm, Maxime Ulysse Garcia, Paolo Di Tommaso & Sven Nahnsen.
Nat Biotechnol. 2020 Feb 13. doi: 10.1038/s41587-020-0439-x.