nf-core/mag
Assembly and binning of metagenomes
1.2.0). The latest
stable release is
5.0.0
.
Introduction
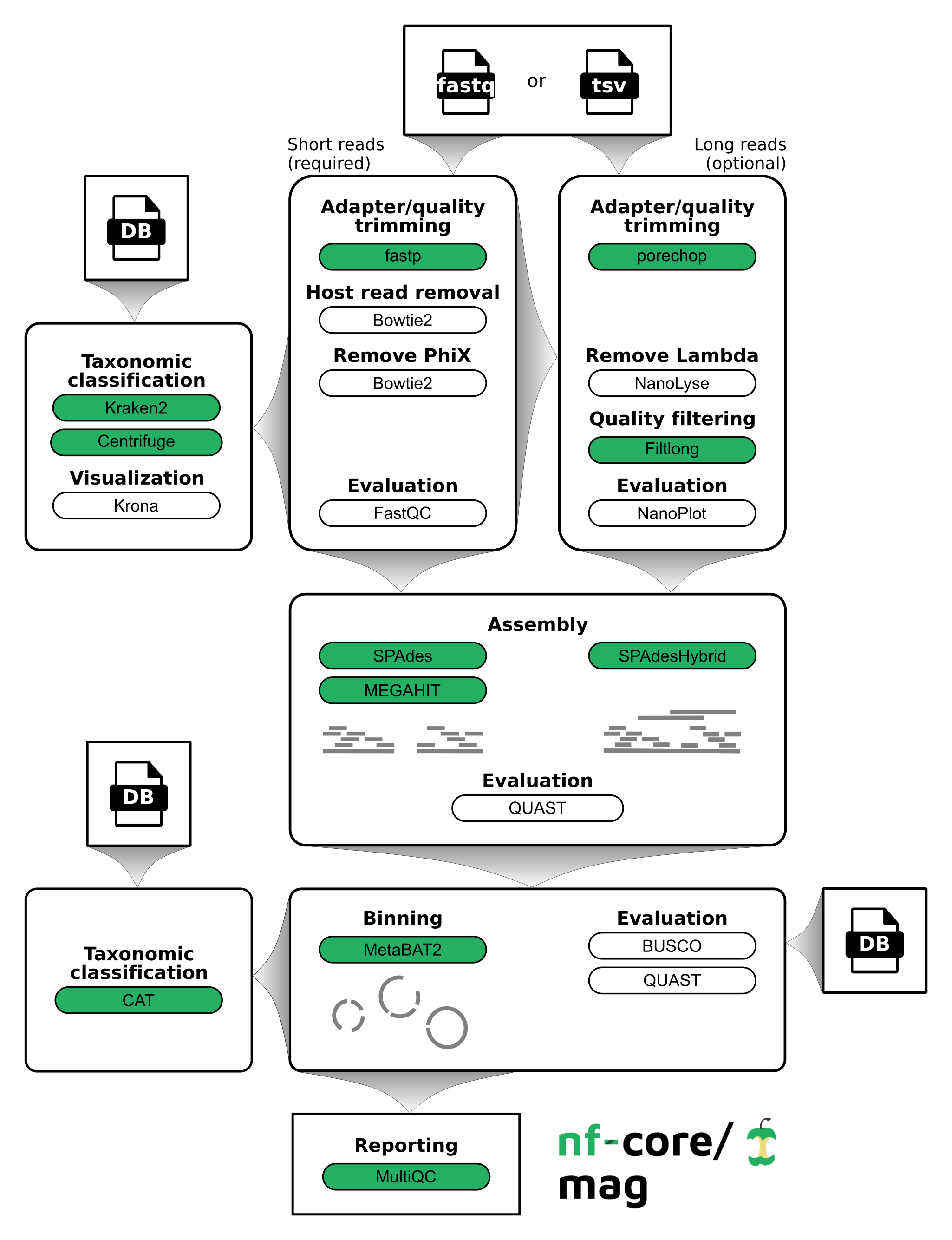
nf-core/mag is a bioinformatics best-practise analysis pipeline for assembly, binning, and annotation of metagenomes.
The pipeline is built using Nextflow, a workflow tool to run tasks across multiple compute infrastructures in a very portable manner. It comes with docker containers making installation trivial and results highly reproducible.
Quick Start
-
Install
nextflow -
Install any of
Docker,SingularityorPodmanfor full pipeline reproducibility (please only useCondaas a last resort; see docs) -
Download the pipeline and test it on a minimal dataset with a single command:
nextflow run nf-core/mag -profile test,<docker/singularity/podman/conda/institute>Please check nf-core/configs to see if a custom config file to run nf-core pipelines already exists for your Institute. If so, you can simply use
-profile <institute>in your command. This will enable eitherdockerorsingularityand set the appropriate execution settings for your local compute environment. -
Start running your own analysis!
nextflow run nf-core/mag -profile <docker/singularity/podman/conda/institute> --input '*_R{1,2}.fastq.gz'or
nextflow run nf-core/mag -profile <docker/singularity/podman/conda/institute> --input 'manifest.tsv'
See usage docs and parameter docs for all of the available options when running the pipeline.
Pipeline Summary
By default, the pipeline currently performs the following: it supports both short and long reads, quality trims the reads and adapters with fastp and Porechop, and performs basic QC with FastQC. The pipeline then:
- assigns taxonomy to reads using Centrifuge and/or Kraken2
- performs assembly using MEGAHIT and SPAdes, and checks their quality using Quast
- performs metagenome binning using MetaBAT2, and checks the quality of the genome bins using Busco
- assigns taxonomy to bins using CAT
Furthermore, the pipeline creates various reports in the results directory specified, including a MultiQC report summarizing some of the findings and software versions.
Documentation
The nf-core/mag pipeline comes with documentation about the pipeline: usage and output. Detailed information about how to specify the input can be found under input specifications.
Group-wise co-assembly and co-abundance computation
Each sample has an associated group ID (see input specifications). This group information can be used for group-wise co-assembly with MEGAHIT or SPAdes and/or to compute co-abundances for the binning step with MetaBAT2. By default, group-wise co-assembly is disabled, while the computation of group-wise co-abundances is enabled. For more information about how this group information can be used see the documentation for the parameters --coassemble_group and --binning_map_mode.
When group-wise co-assembly is enabled, SPAdes is run on accordingly pooled read files, since metaSPAdes does not yet allow the input of multiple samples or libraries. In contrast, MEGAHIT is run for each group while supplying lists of the individual readfiles.
Credits
nf-core/mag was written by Hadrien Gourlé at SLU, Daniel Straub and Sabrina Krakau at the Quantitative Biology Center (QBiC).
Long read processing was inspired by caspargross/HybridAssembly written by Caspar Gross @caspargross
Many thanks to the additional contributors who have helped out and/or provided suggestions:
- Alexander Peltzer
- Phil Ewels
- Gisela Gabernet
- Harshil Patel
- Johannes Alneberg
- Maxime Borry
- Maxime Garcia
- Michael L Heuer
Contributions and Support
If you would like to contribute to this pipeline, please see the contributing guidelines.
For further information or help, don’t hesitate to get in touch on the Slack #mag channel (you can join with this invite).
Citations
If you use nf-core/mag for your analysis, please cite it using the following doi: 10.5281/zenodo.3589527
You can cite the nf-core publication as follows:
The nf-core framework for community-curated bioinformatics pipelines.
Philip Ewels, Alexander Peltzer, Sven Fillinger, Harshil Patel, Johannes Alneberg, Andreas Wilm, Maxime Ulysse Garcia, Paolo Di Tommaso & Sven Nahnsen.
Nat Biotechnol. 2020 Feb 13. doi: 10.1038/s41587-020-0439-x. ReadCube: Full Access Link
References of tools used in this pipeline are as follows:
- Bowtie2 Langmead, B. and Salzberg, S. L. 2012 Fast gapped-read alignment with Bowtie 2. Nature methods, 9(4), p. 357–359. doi: 10.1038/nmeth.1923.
- Busco Seppey, M., Manni, M., & Zdobnov, E. M. (2019). BUSCO: assessing genome assembly and annotation completeness. In Gene prediction (pp. 227-245). Humana, New York, NY. https://doi.org/10.1007/978-1-4939-9173-0_14
- CAT von Meijenfeldt, F. B., Arkhipova, K., Cambuy, D. D., Coutinho, F. H., & Dutilh, B. E. (2019). Robust taxonomic classification of uncharted microbial sequences and bins with CAT and BAT. Genome biology, 20(1), 1-14. https://doi.org/10.1186/s13059-019-1817-x. Home: https://github.com/dutilh/CAT
- Centrifuge Kim, D., Song, L., Breitwieser, F. P., & Salzberg, S. L. (2016). Centrifuge: rapid and sensitive classification of metagenomic sequences. Genome research, 26(12), 1721-1729. https://doi.org/10.1101/gr.210641.116. Home: http://www.ccb.jhu.edu/software/centrifuge
- FastP Chen, S., Zhou, Y., Chen, Y., & Gu, J. (2018). fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics , 34(17), i884–i890. https://doi.org/10.1093/bioinformatics/bty560. Home: https://github.com/OpenGene/fastp
- FastQC Home: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/
- Filtlong Home: https://github.com/rrwick/Filtlong
- Kraken2 Wood, D et al., 2019. Improved metagenomic analysis with Kraken 2. Genome Biology volume 20, Article number: 257. https://doi.org/10.1186/s13059-019-1891-0. Home: https://ccb.jhu.edu/software/kraken2/
- Krona Ondov, B. D., Bergman, N. H., & Phillippy, A. M. (2011). Interactive metagenomic visualization in a Web browser. BMC bioinformatics, 12(1), 1-10. https://doi.org/10.1186/1471-2105-12-385. Home: https://github.com/marbl/Krona/wiki.
- MEGAHIT Li, D., Luo, R., Liu, C. M., Leung, C. M., Ting, H. F., Sadakane, K., … & Lam, T. W. (2016). MEGAHIT v1. 0: a fast and scalable metagenome assembler driven by advanced methodologies and community practices. Methods, 102, 3-11. https://doi.org/10.1016/j.ymeth.2016.02.020. Home: https://github.com/voutcn/megahit
- MetaBAT2 Kang, D. D., Li, F., Kirton, E., Thomas, A., Egan, R., An, H., & Wang, Z. (2019). MetaBAT 2: an adaptive binning algorithm for robust and efficient genome reconstruction from metagenome assemblies. PeerJ, 7, e7359. https://doi.org/10.7717/peerj.7359. Home: https://bitbucket.org/berkeleylab/metabat
- MultiQC Ewels, P., Magnusson, M., Lundin, S., & Käller, M. (2016). MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics , 32(19), 3047–3048. https://doi.org/10.1093/bioinformatics/btw354. Home: https://multiqc.info/
- NanoLyse De Coster, W., D’Hert, S., Schultz, D. T., Cruts, M., & Van Broeckhoven, C. (2018). NanoPack: visualizing and processing long-read sequencing data. Bioinformatics, 34(15), 2666-2669. https://doi.org/10.1093/bioinformatics/bty149. Home: https://github.com/wdecoster/nanolyse
- NanoPlot De Coster, W., D’Hert, S., Schultz, D. T., Cruts, M., & Van Broeckhoven, C. (2018). NanoPack: visualizing and processing long-read sequencing data. Bioinformatics, 34(15), 2666-2669. https://doi.org/10.1093/bioinformatics/bty149. Home: https://github.com/wdecoster/NanoPlot
- Porechop Home: https://github.com/rrwick/Porechop
- SAMtools Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., … 1000 Genome Project Data Processing Subgroup. (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics , 25(16), 2078–2079. https://doi.org/10.1093/bioinformatics/btp352. Home: http://www.htslib.org/
- SPAdes Nurk, S., Meleshko, D., Korobeynikov, A., & Pevzner, P. A. (2017). metaSPAdes: a new versatile metagenomic assembler. Genome research, 27(5), 824-834. https://doi.org/10.1101/gr.213959.116
In addition, this repository uses test data from the following study:
- Bertrand, D., Shaw, J., Kalathiyappan, M., Ng, A. H. Q., Kumar, M. S., Li, C., … & Nagarajan, N. (2019). Hybrid metagenomic assembly enables high-resolution analysis of resistance determinants and mobile elements in human microbiomes. Nature biotechnology, 37(8), 937-944. https://doi.org/10.1038/s41587-019-0191-2